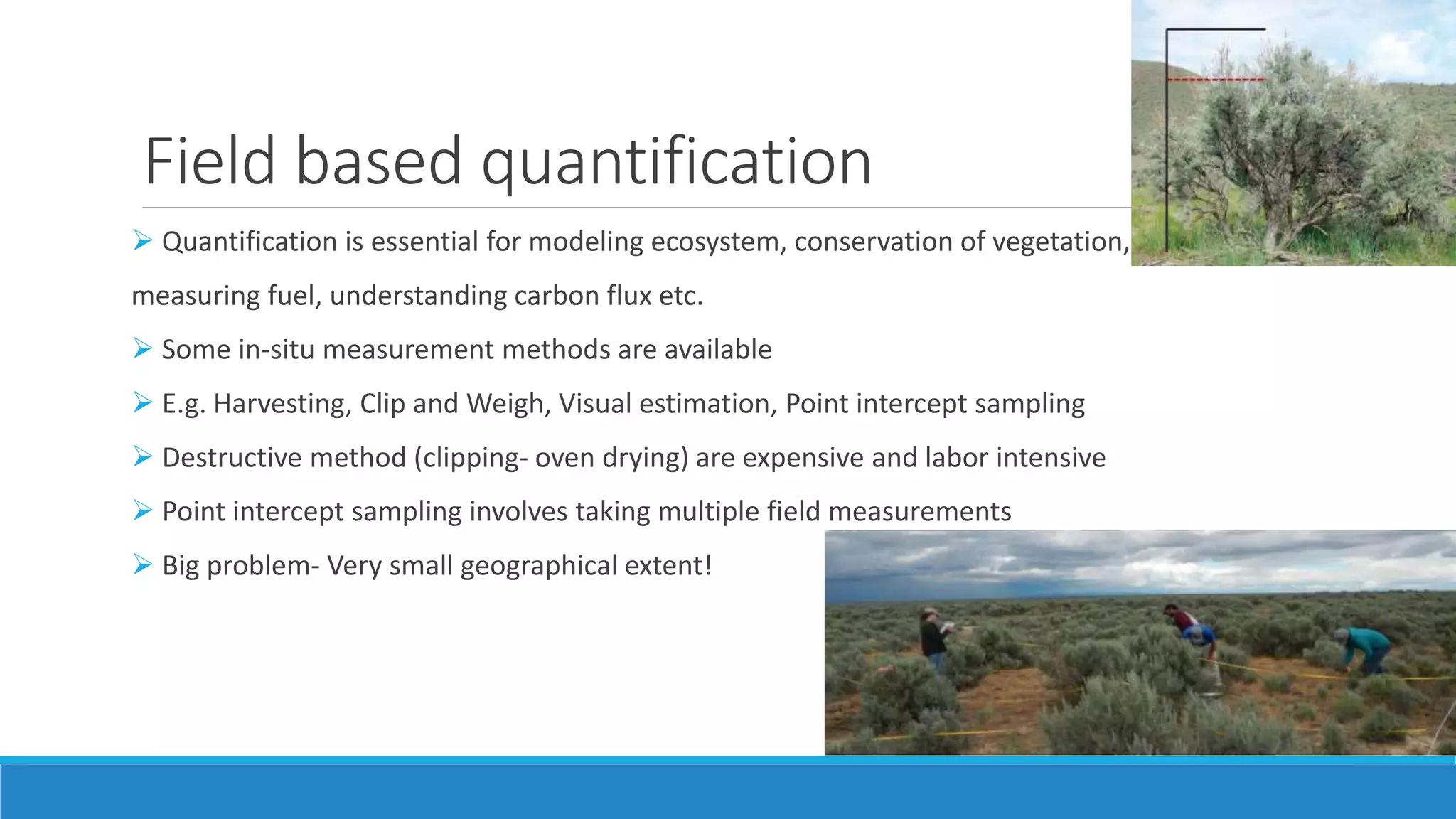
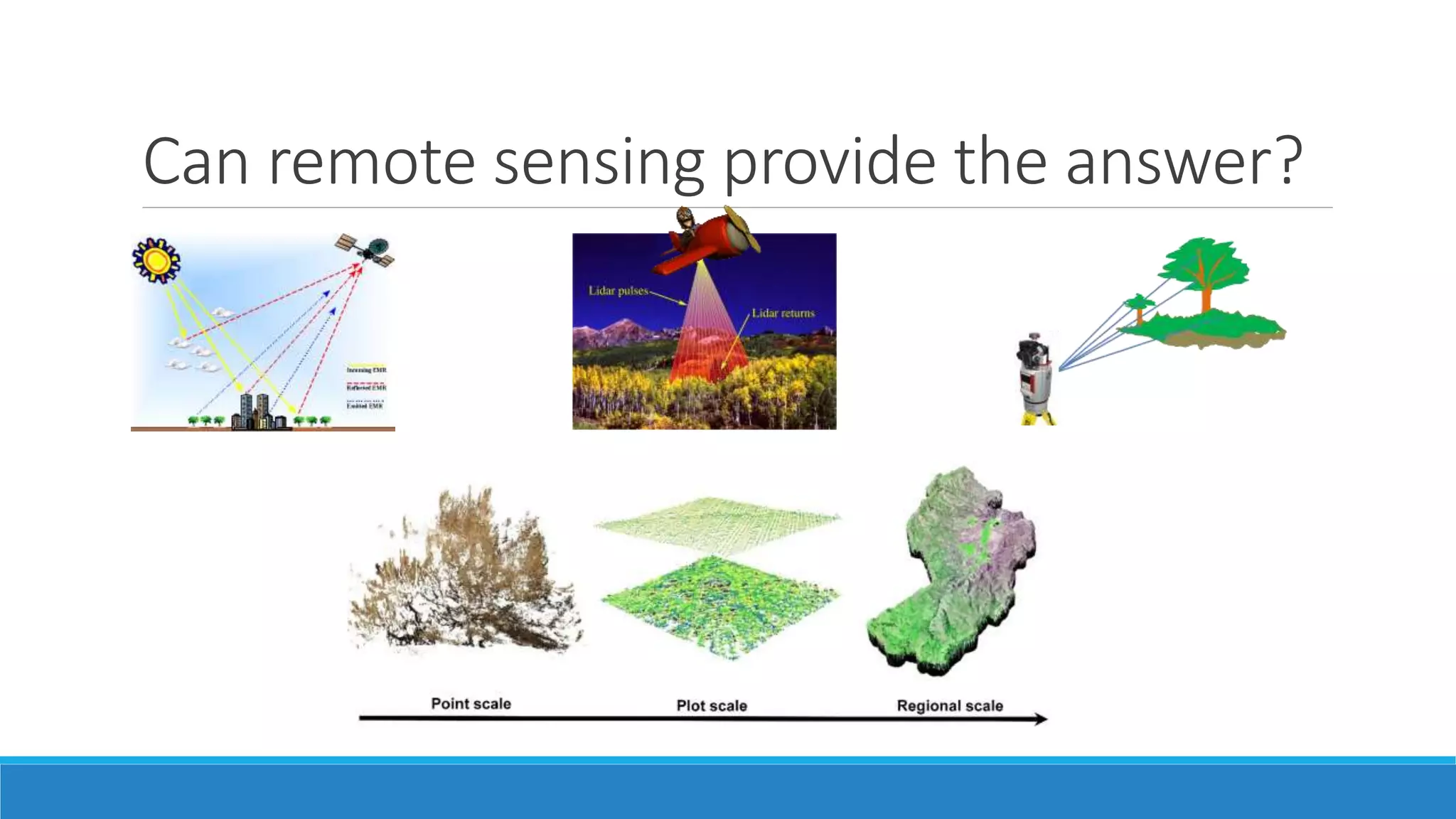
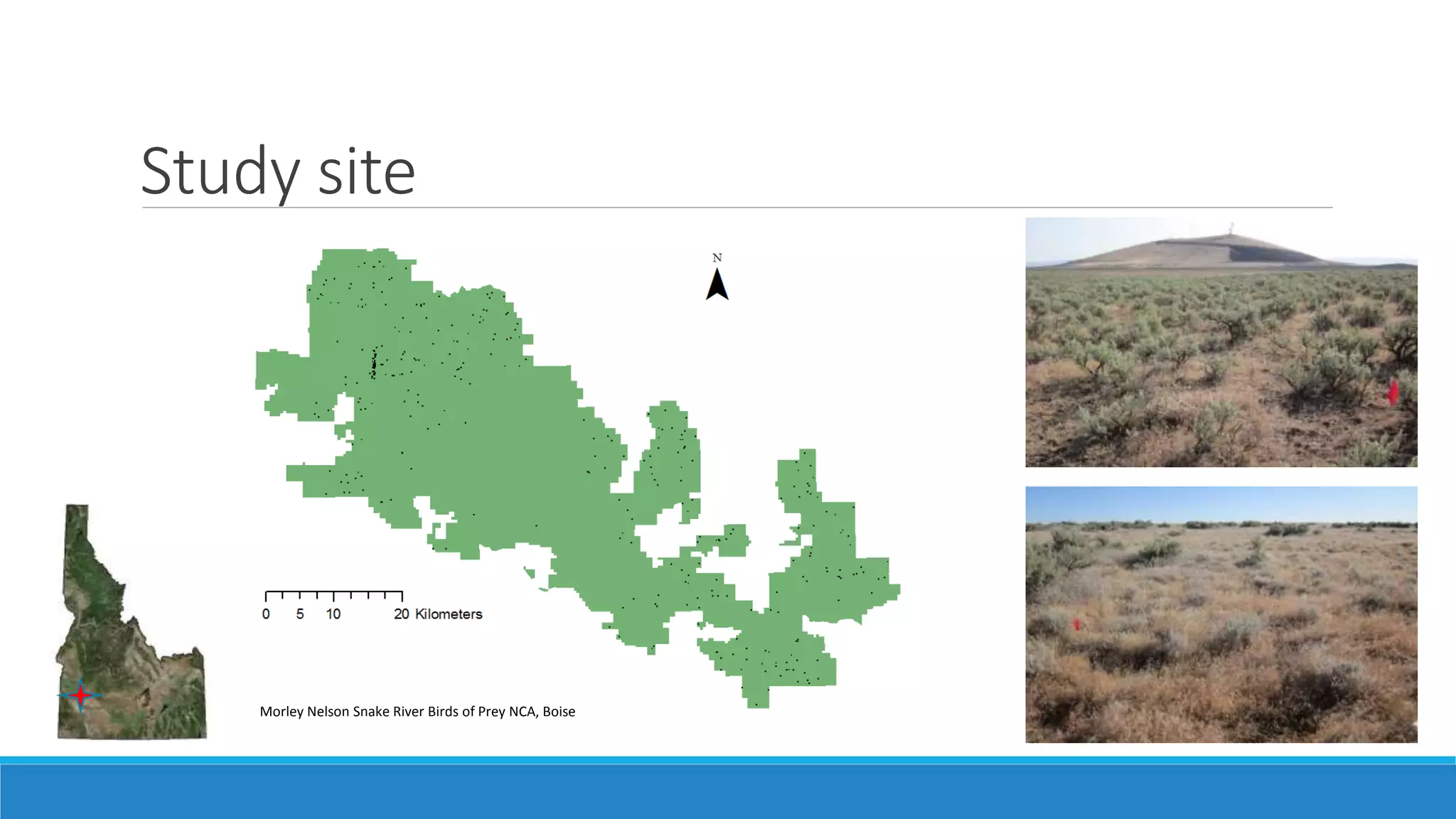
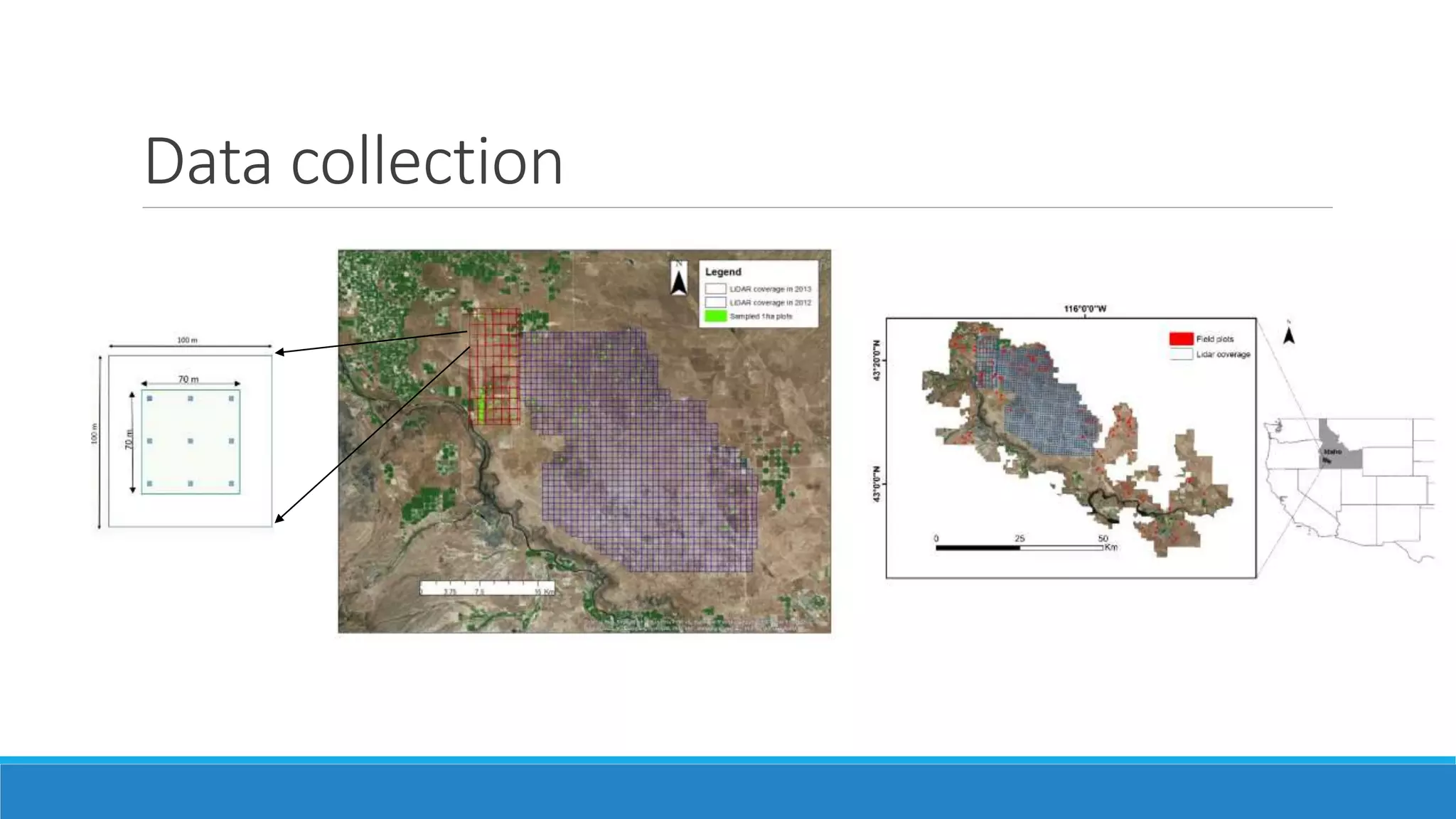
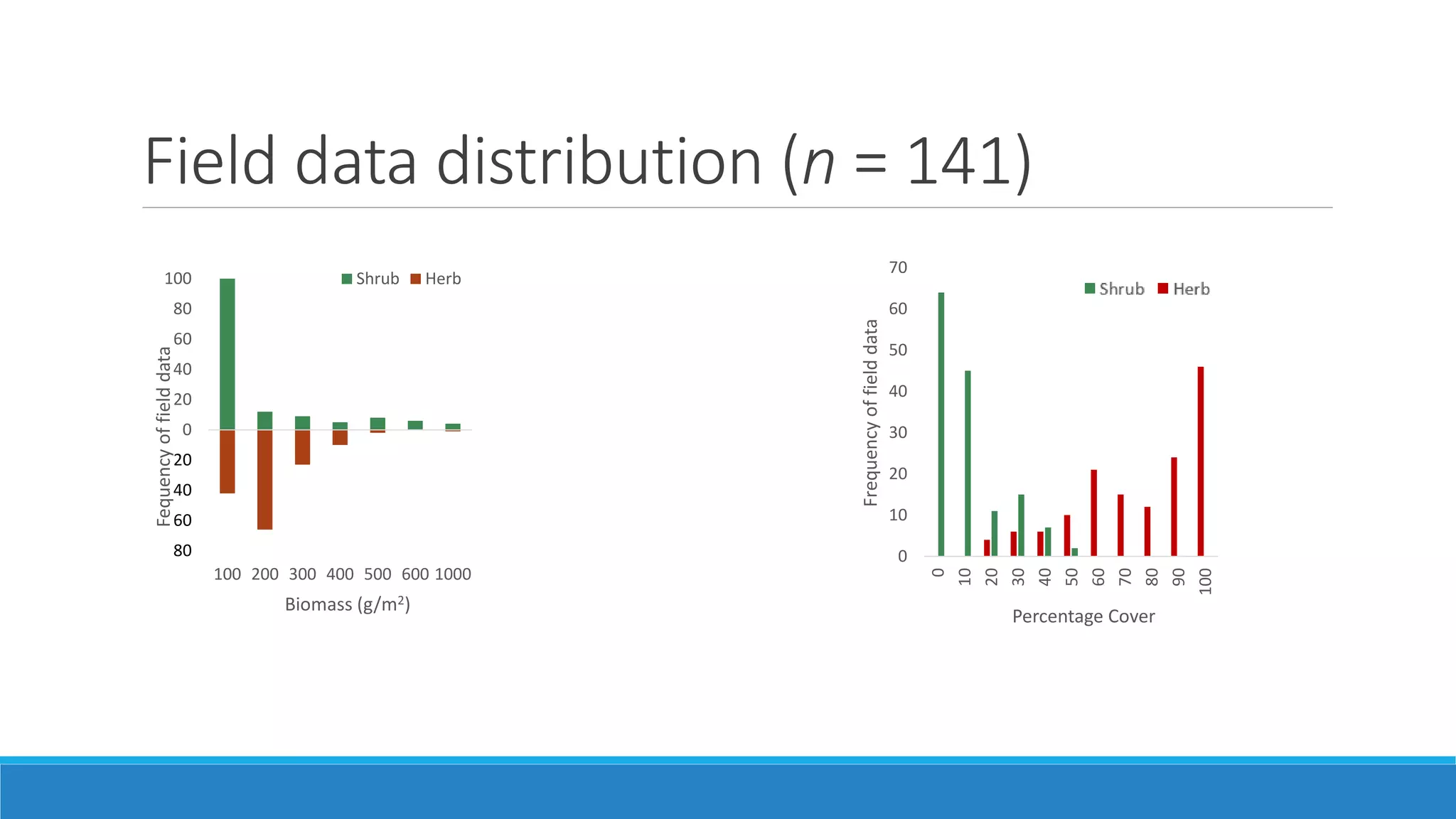
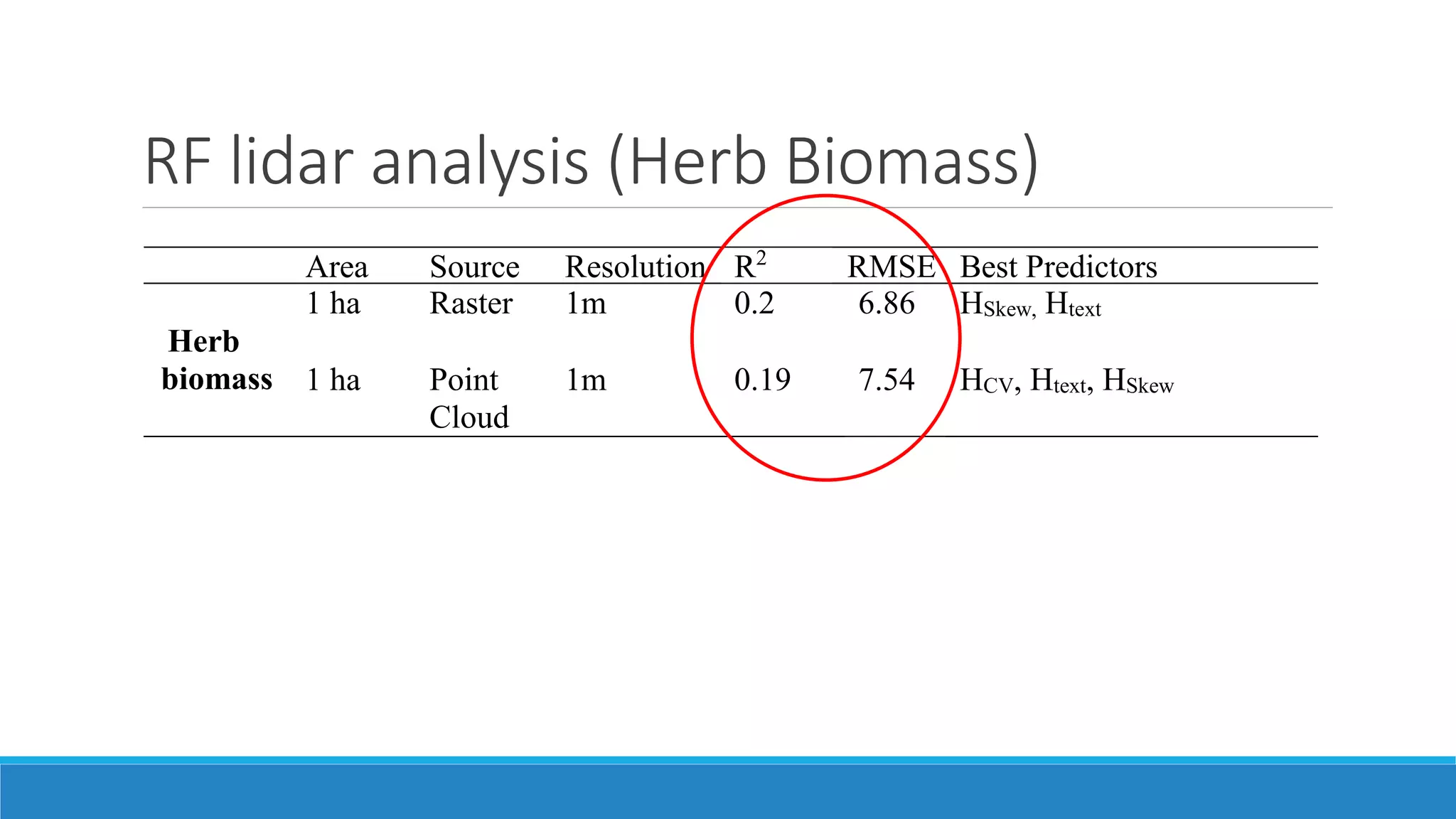
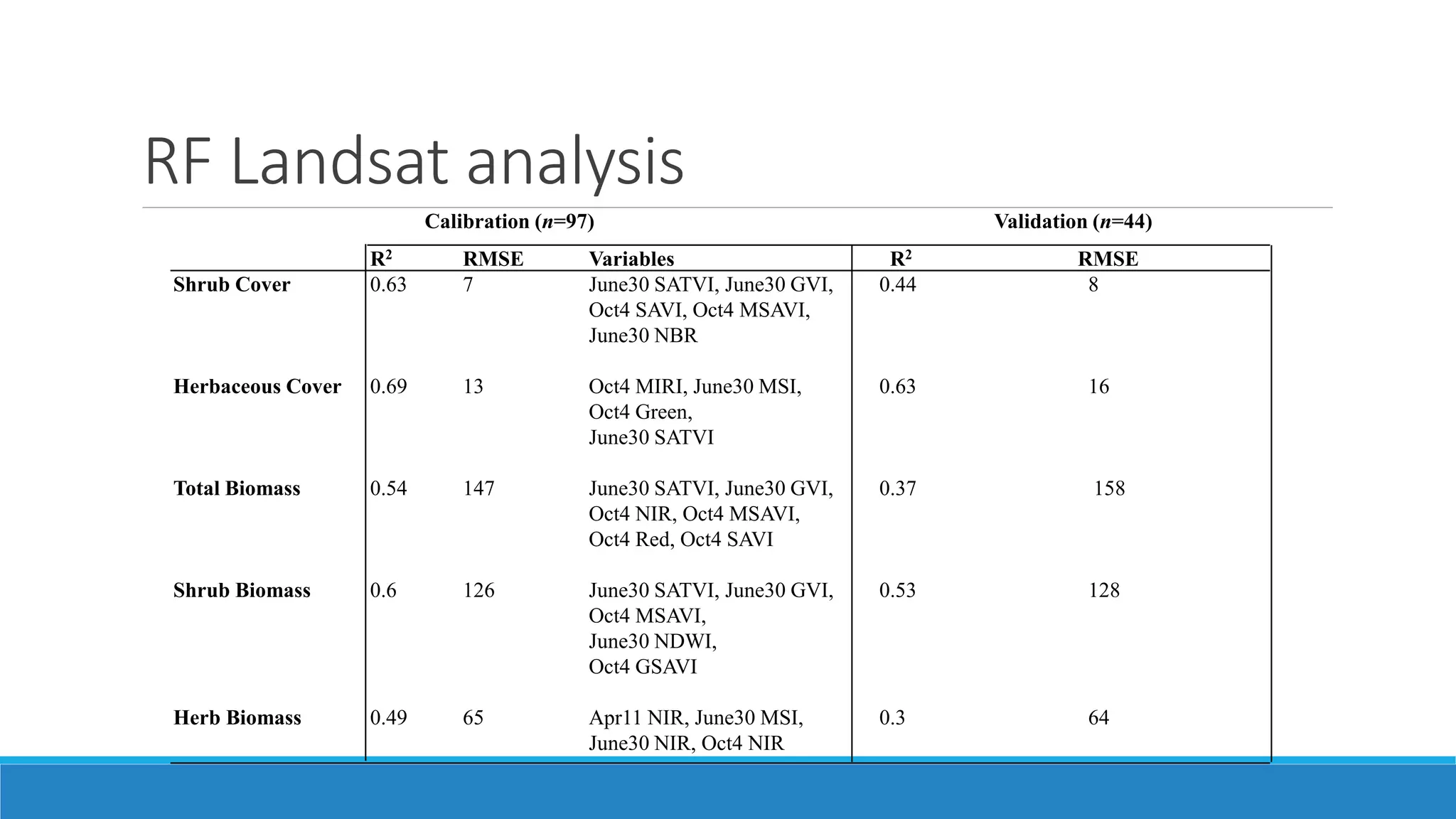
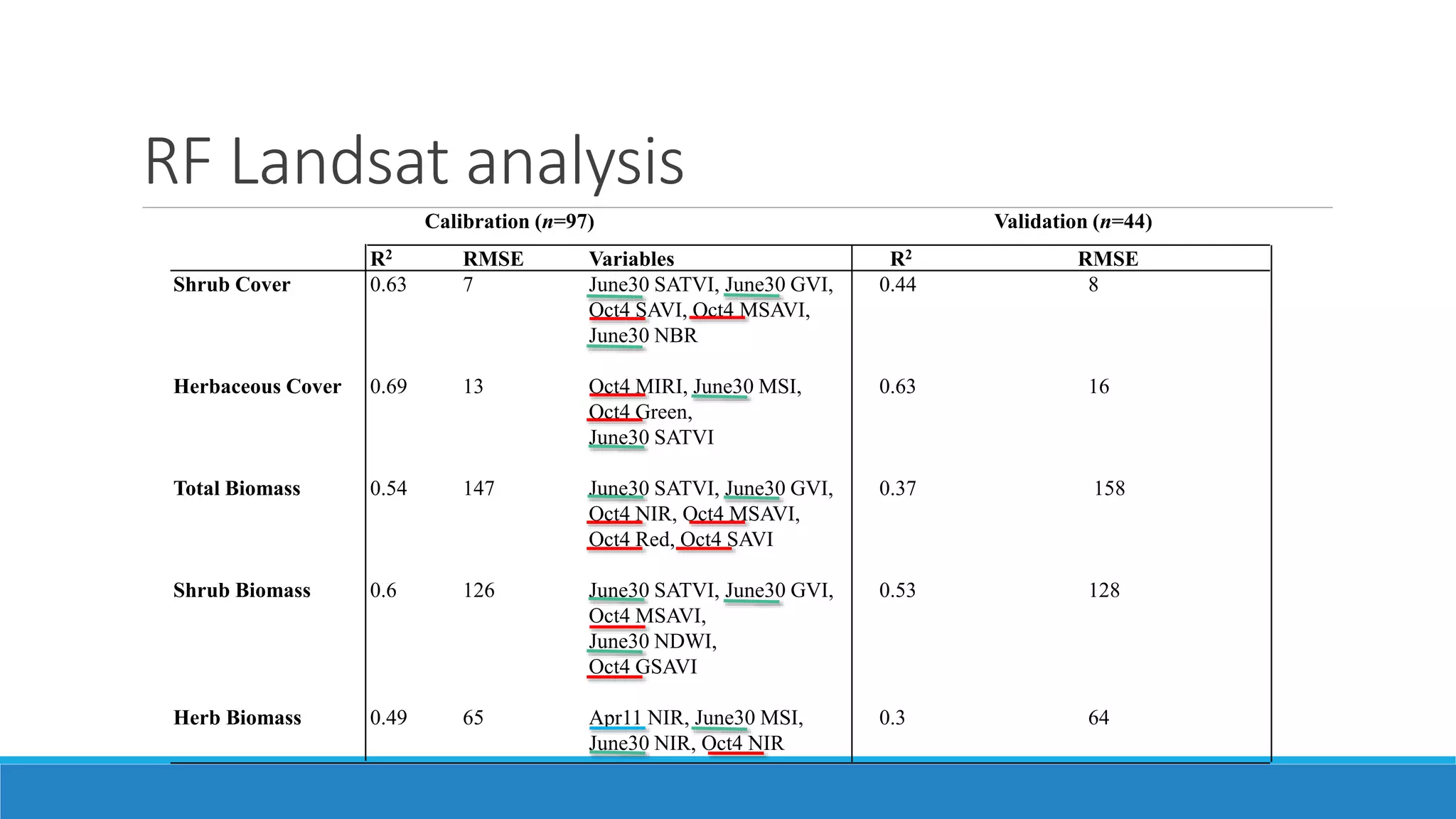
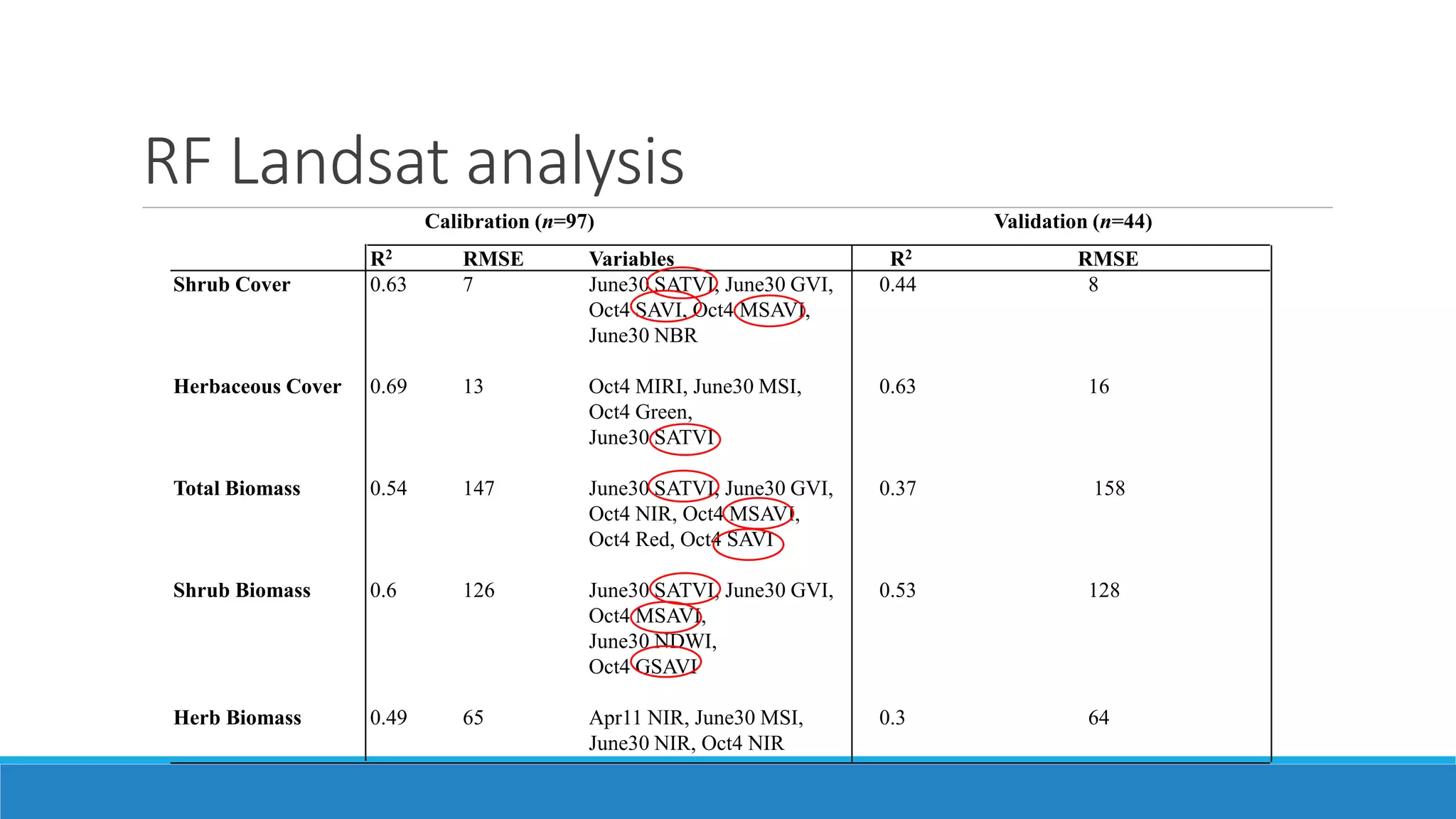
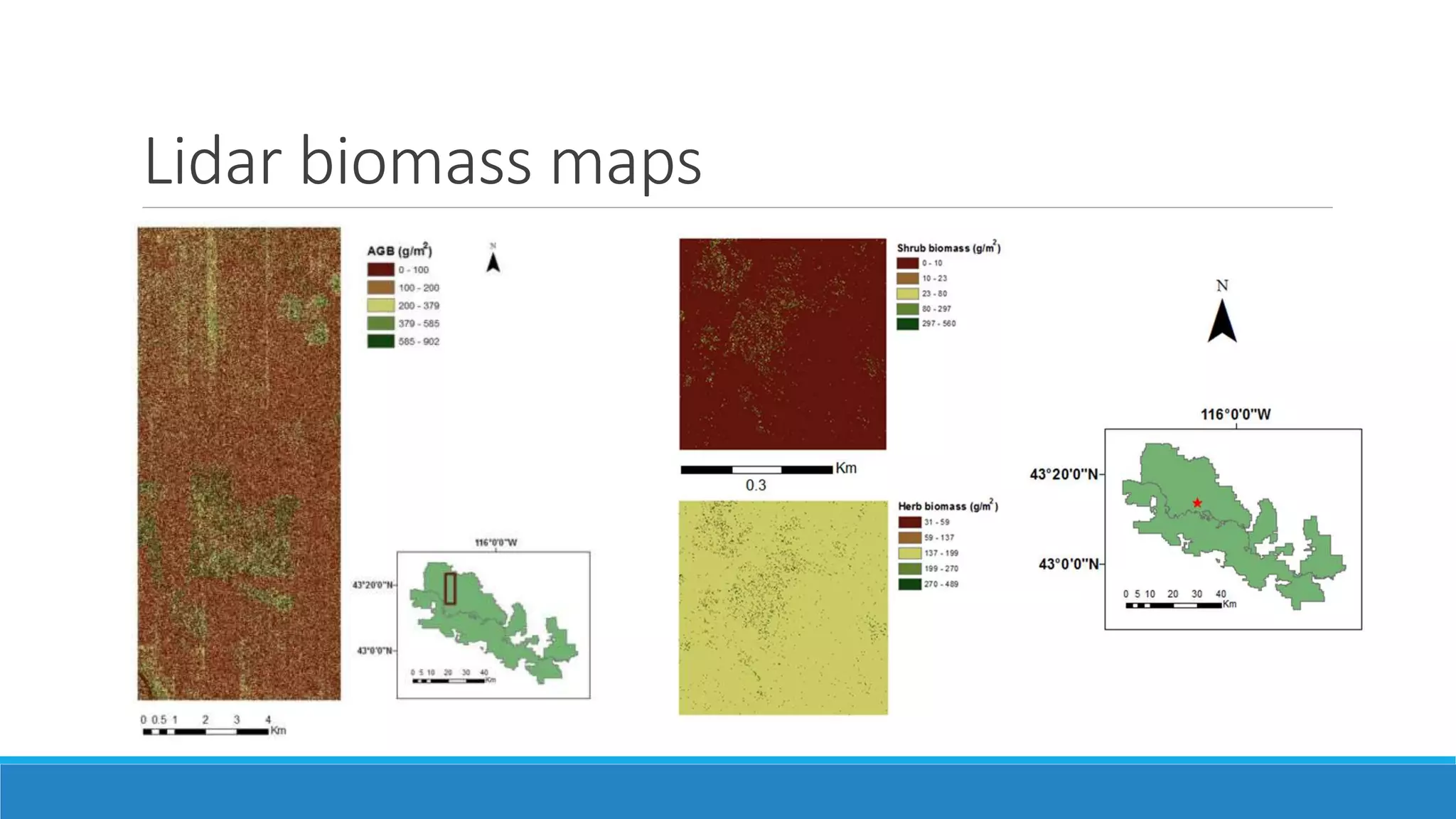
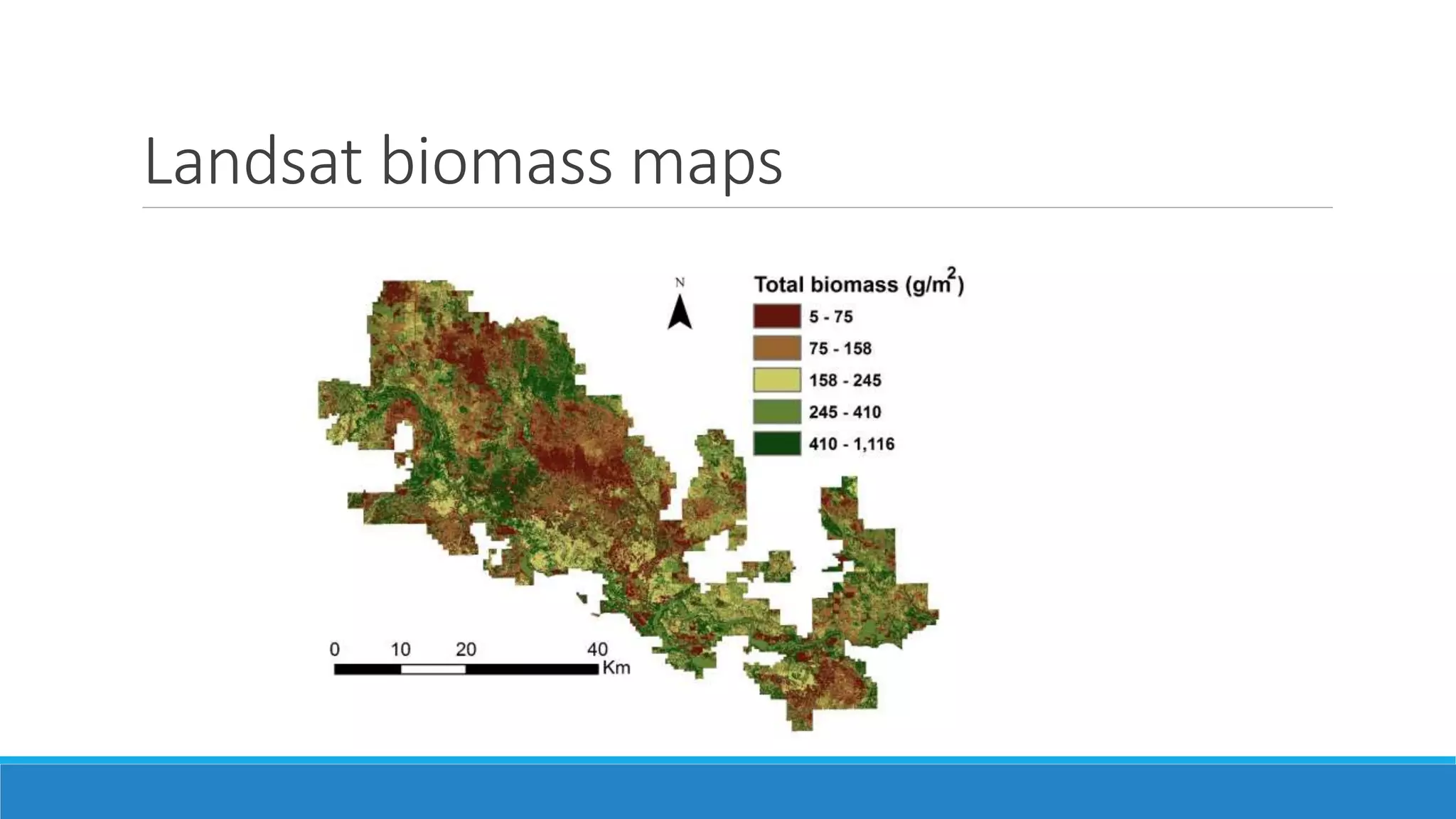
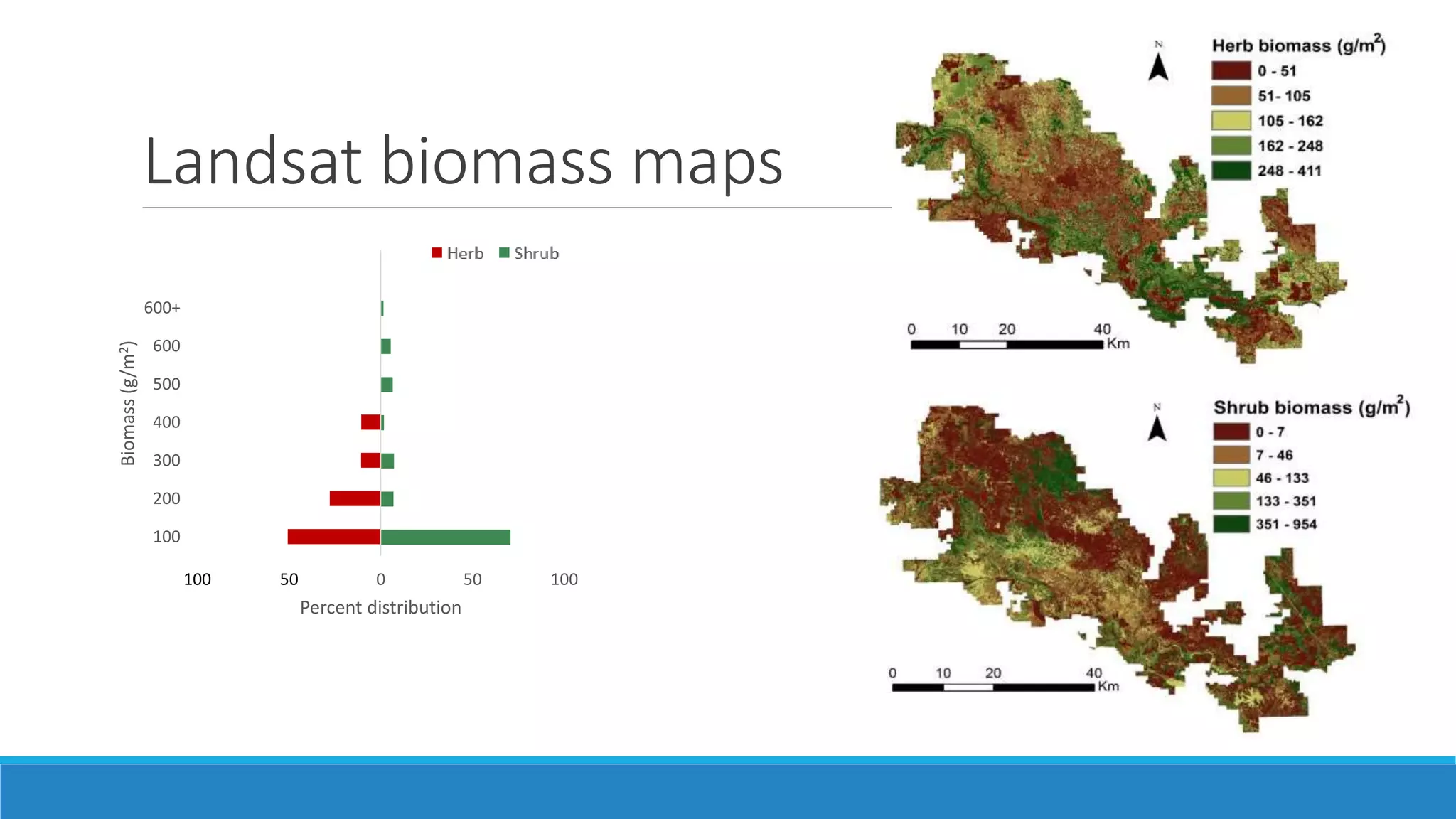
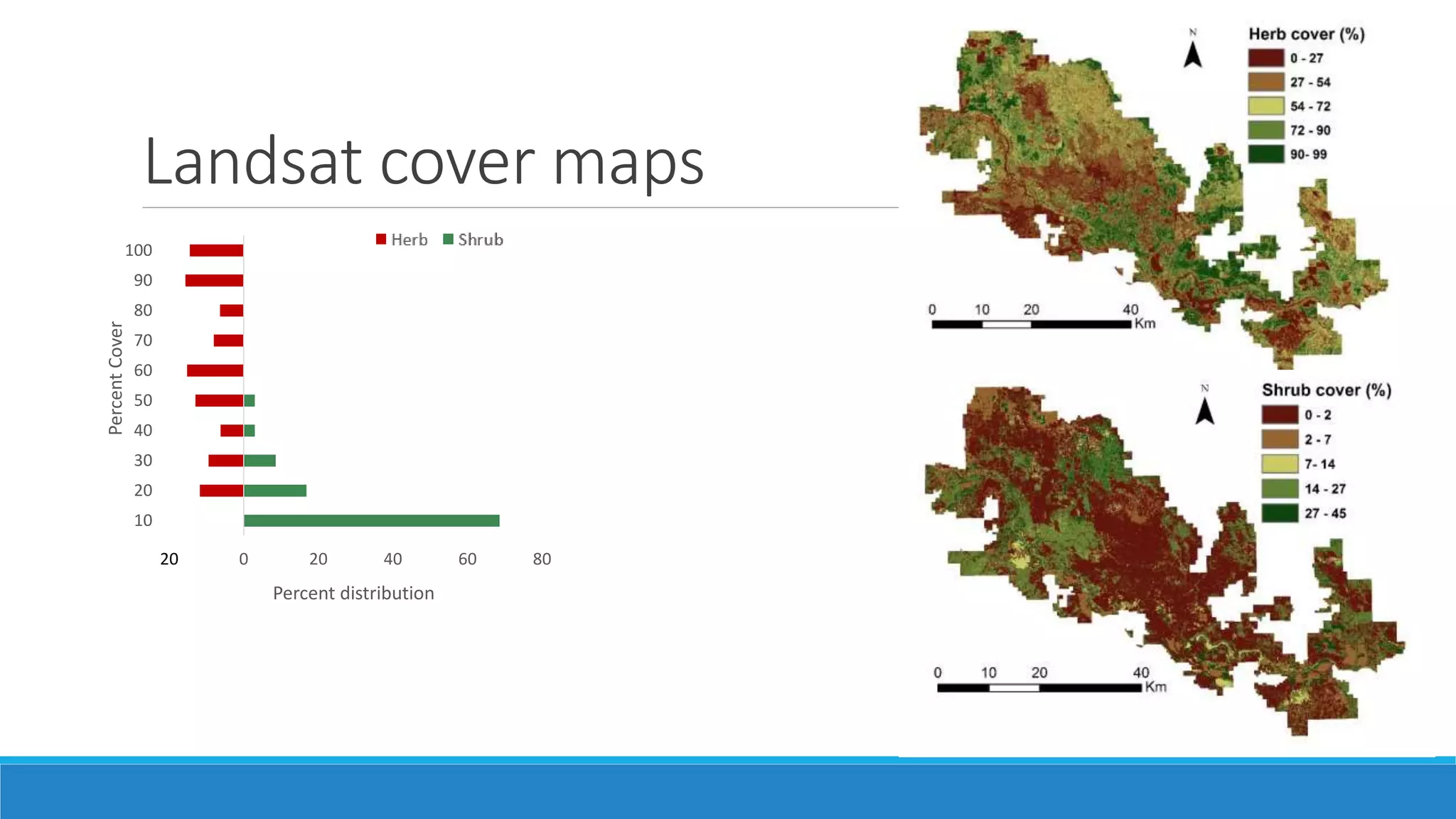
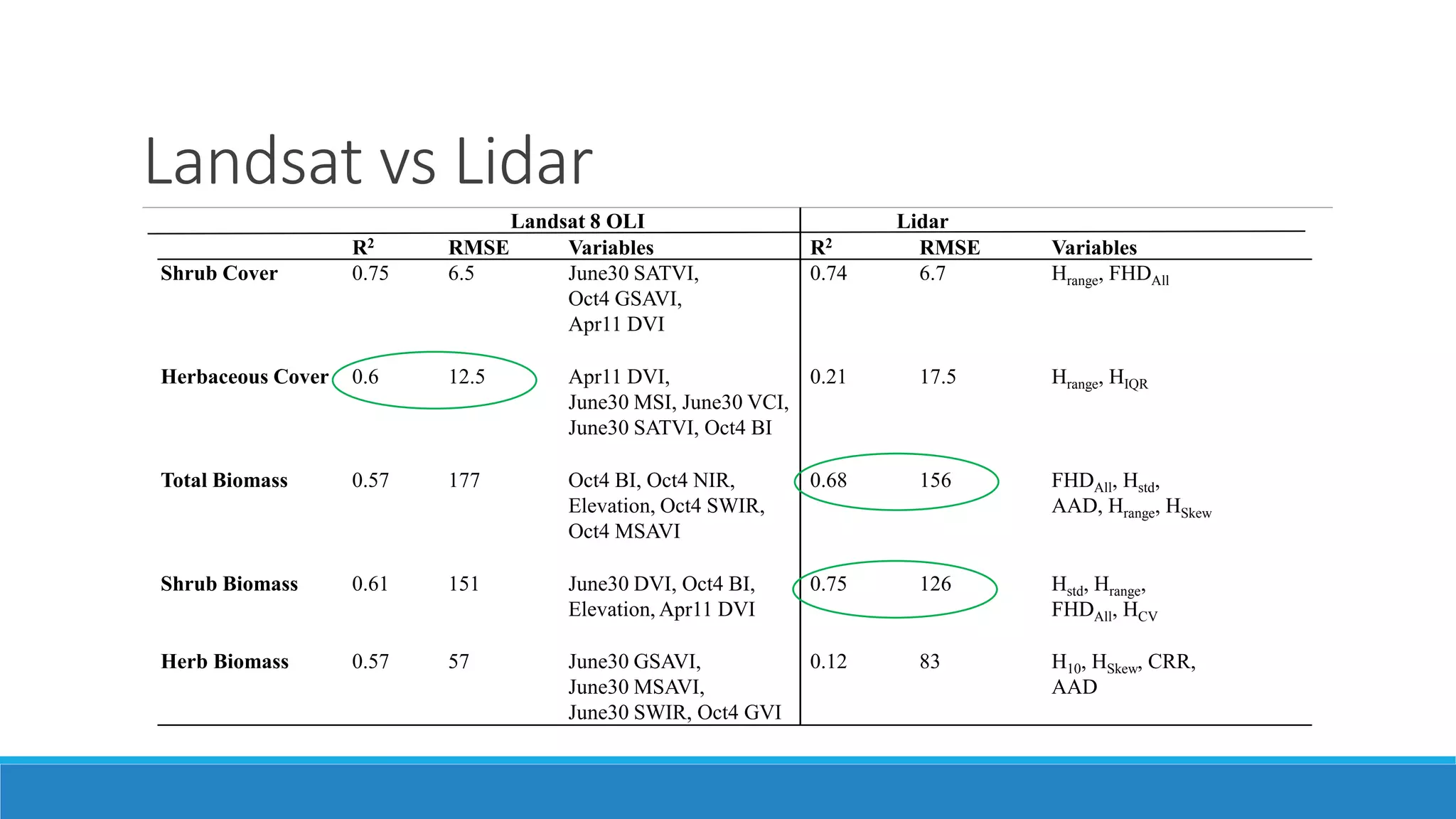
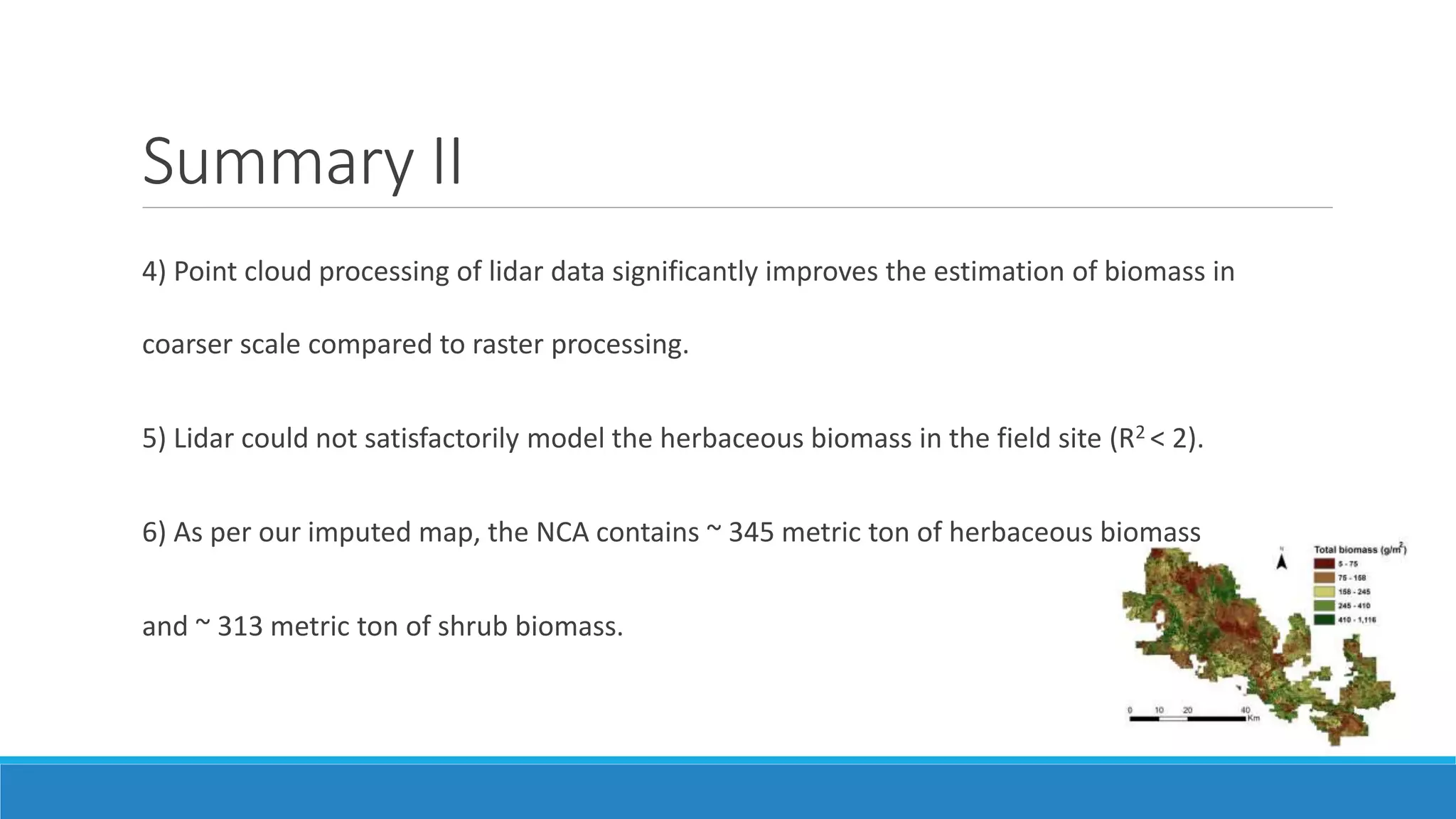
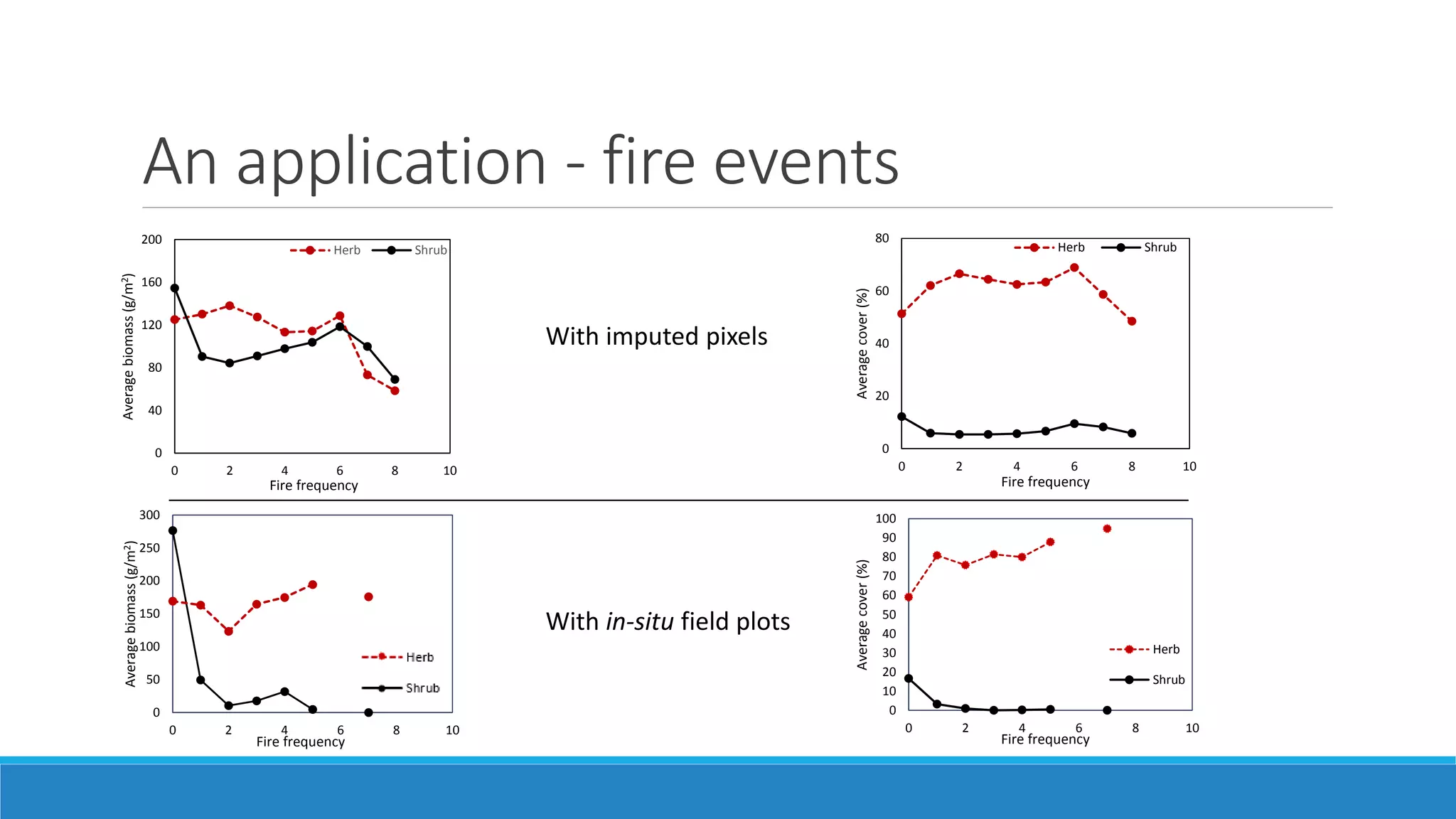
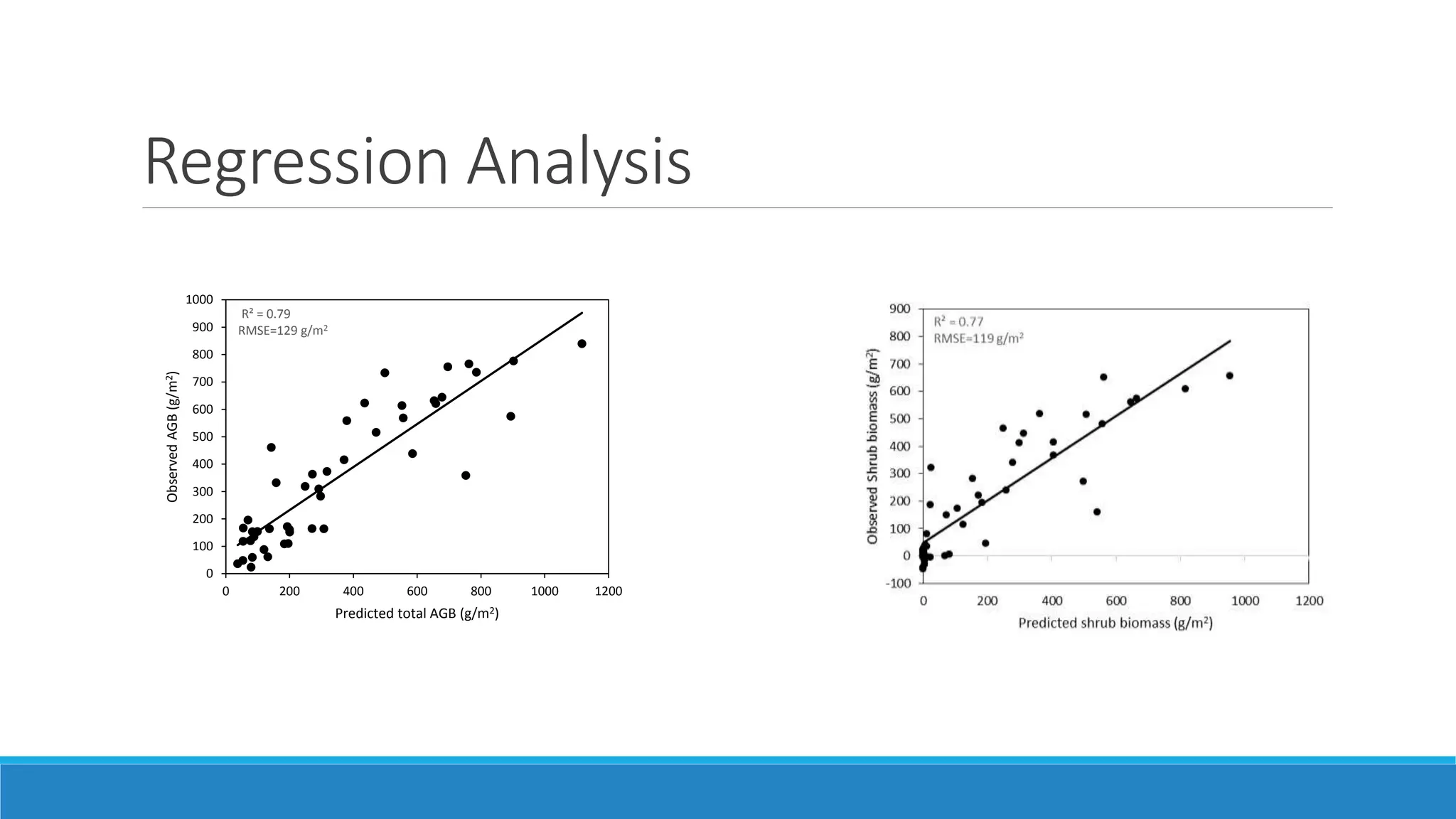
This document evaluates the effectiveness of lidar and Landsat for estimating aboveground vegetation biomass and cover in semi-arid rangelands through machine learning algorithms. The study finds that lidar is superior for estimating shrub biomass, while Landsat is suitable for broader herbaceous cover estimations, suggesting a synergistic use of both technologies for better mapping and modeling. Recommendations for practical applications include aiding land managers and ecologists in large-scale vegetation assessments and potential global adaptation of methods.