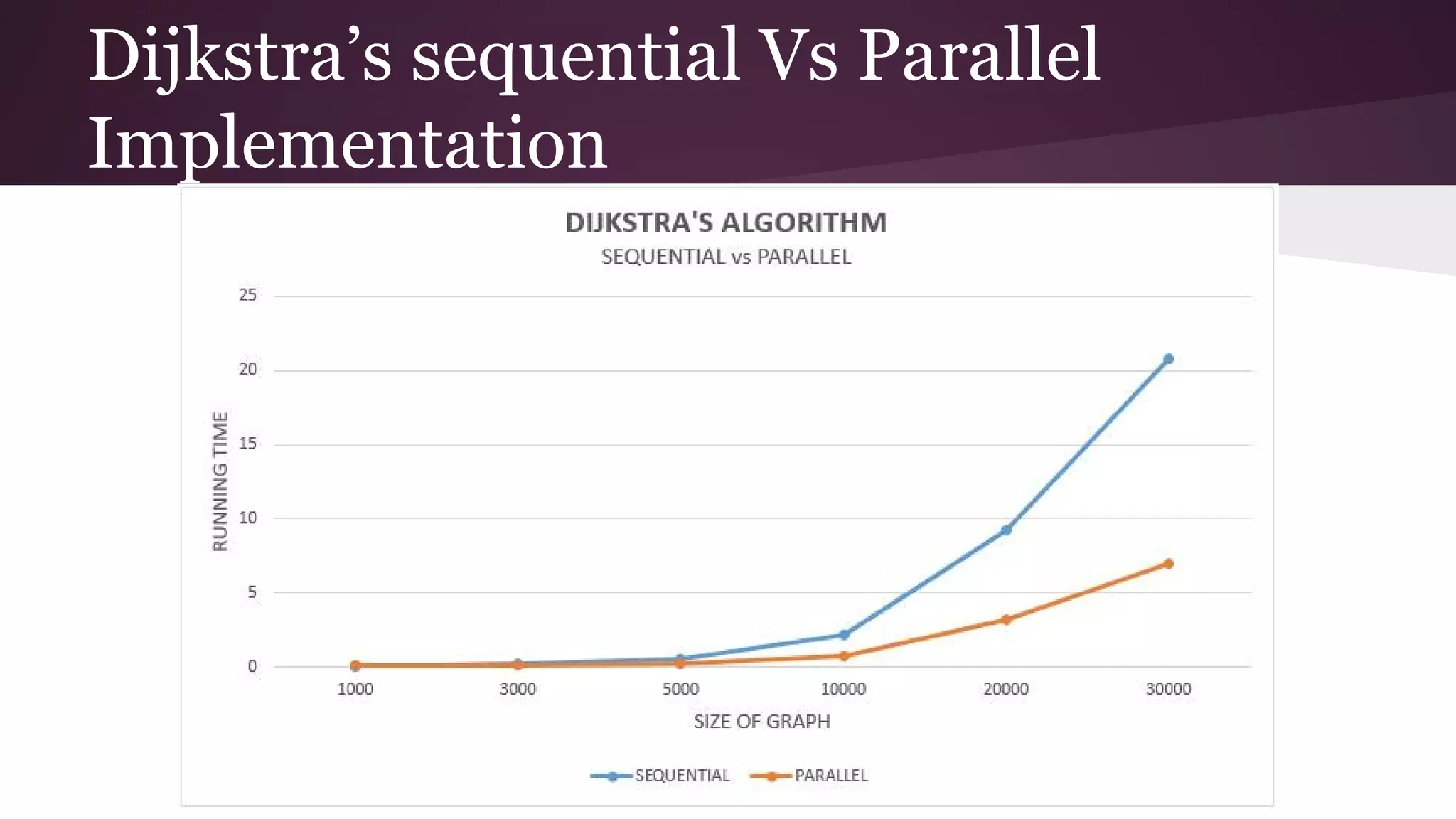
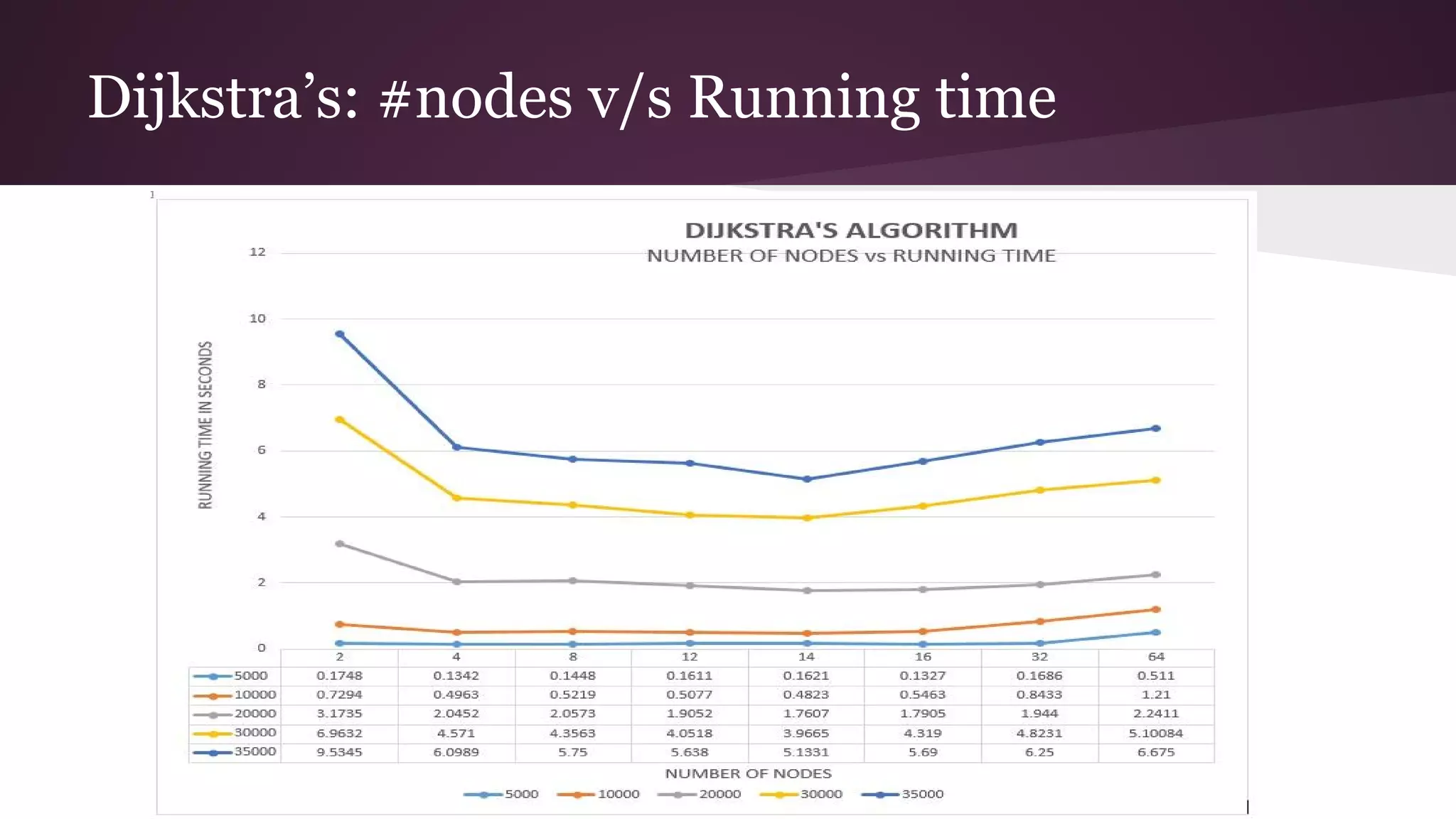
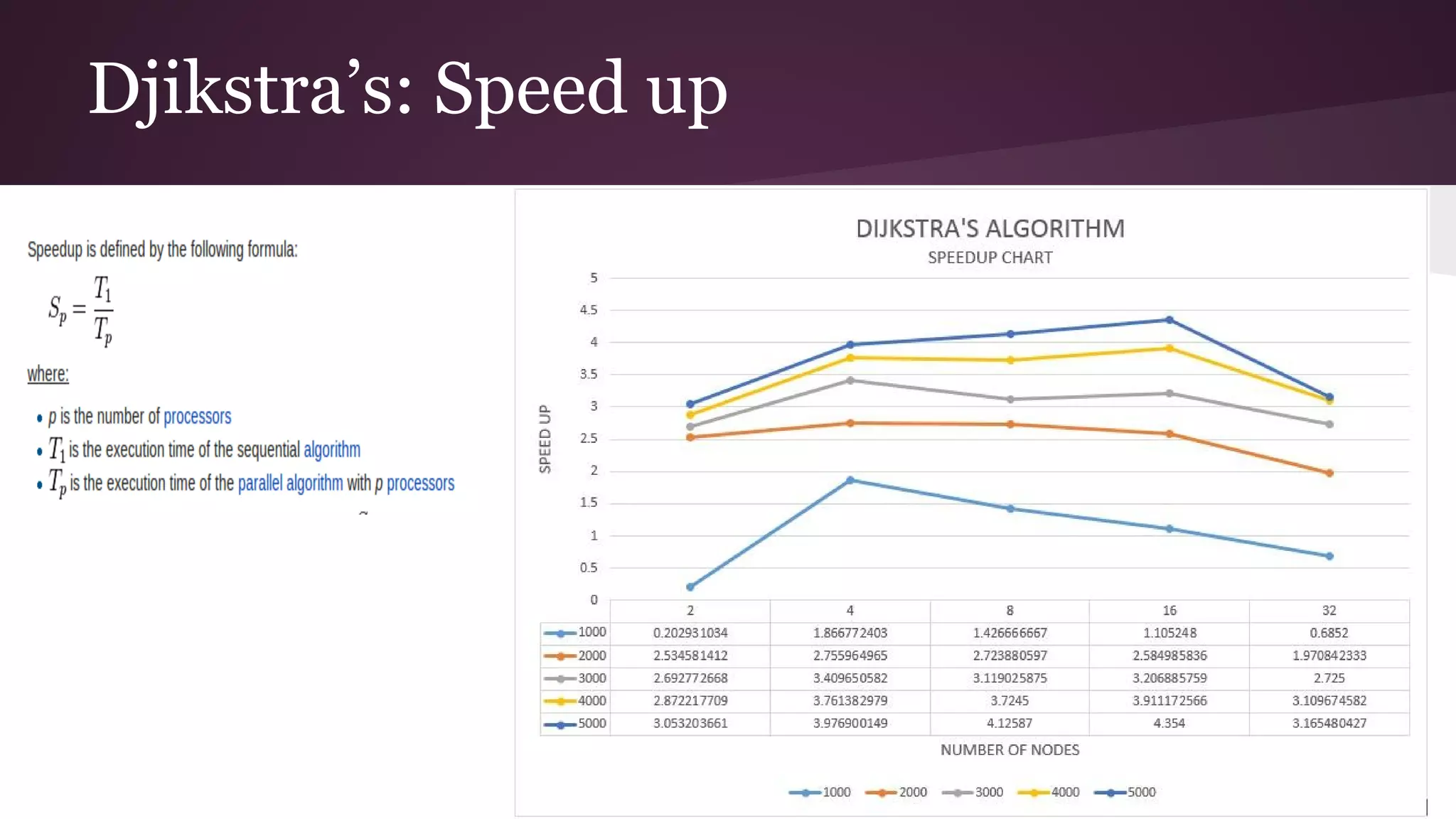
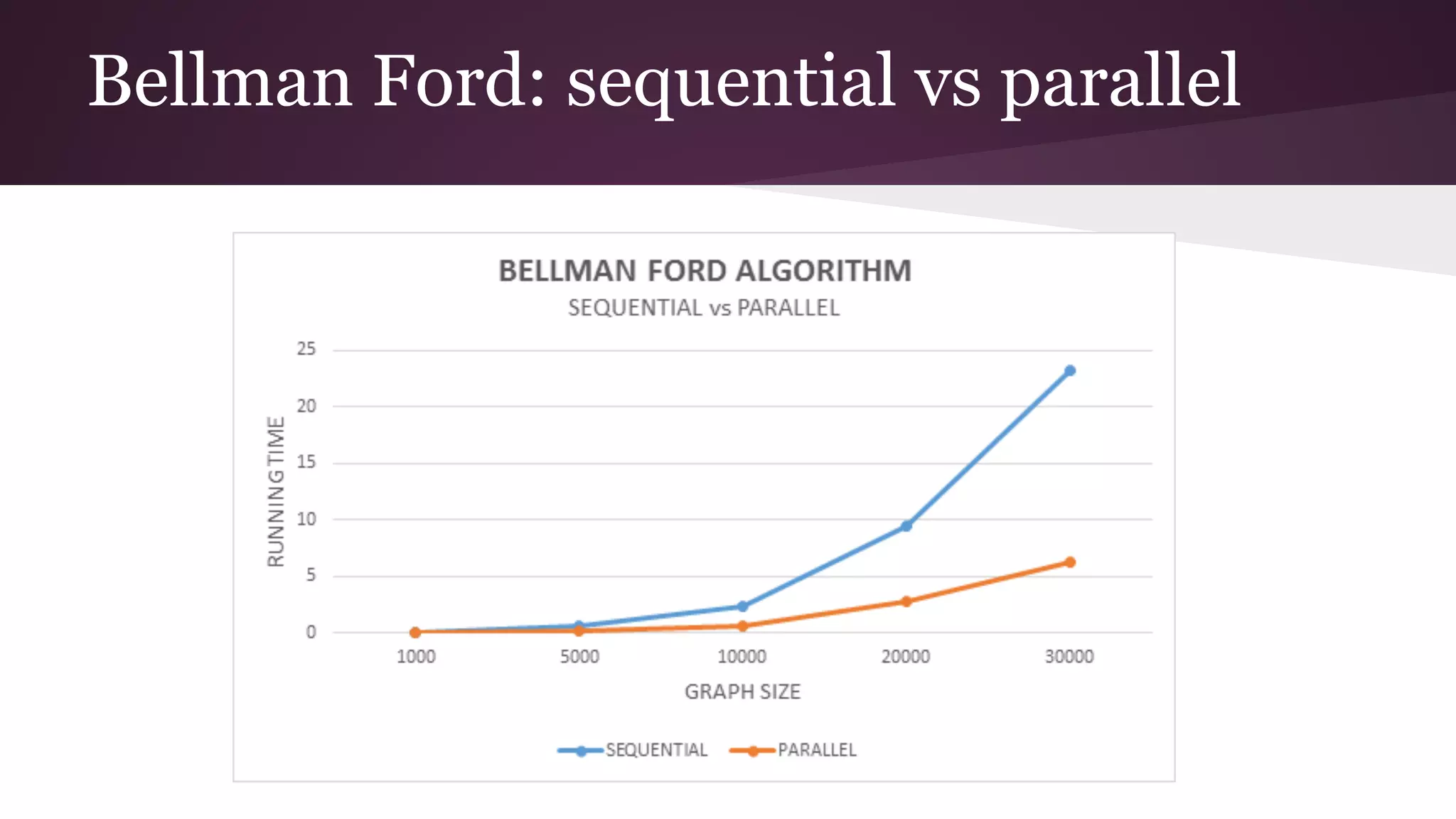
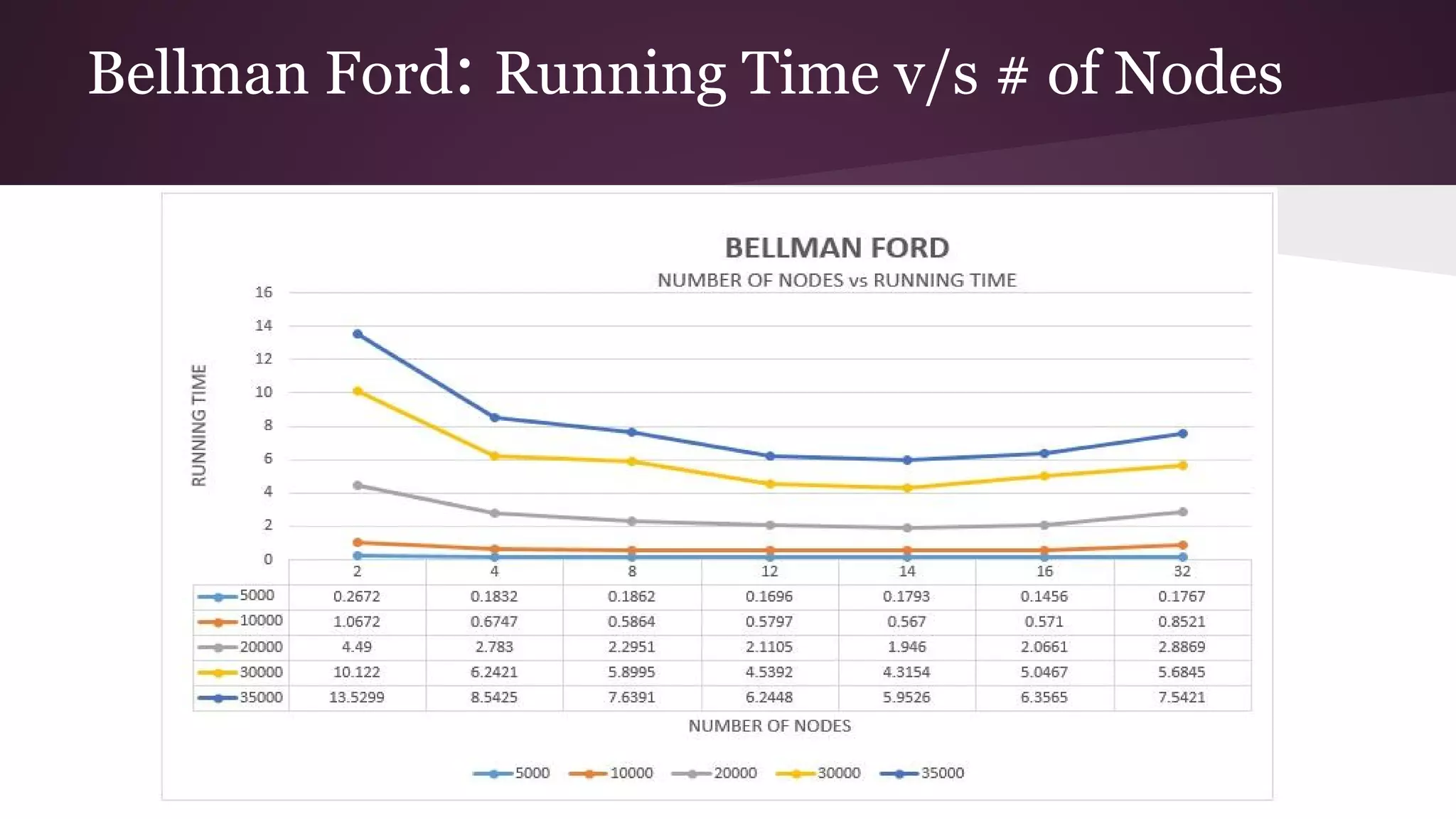
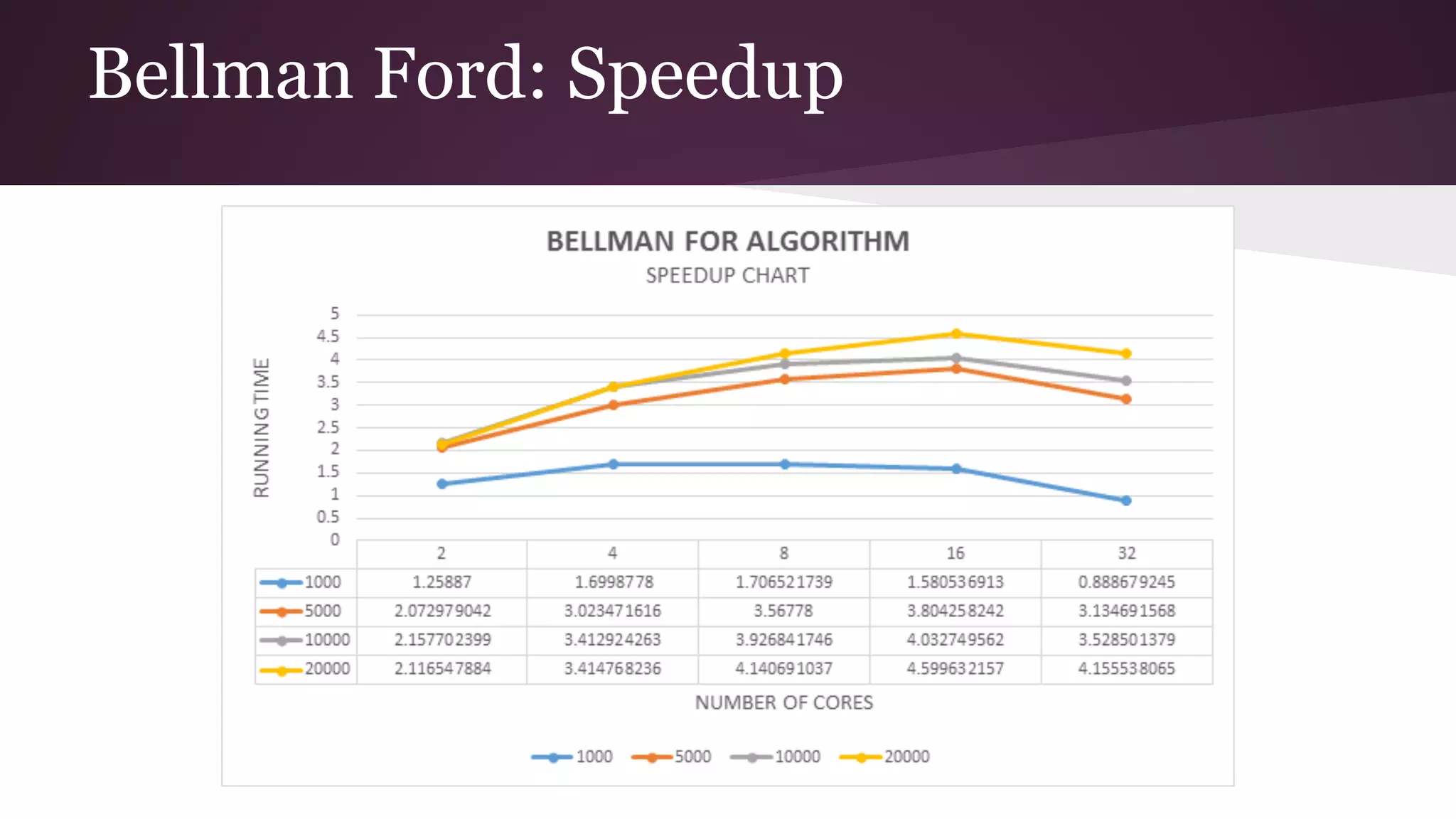
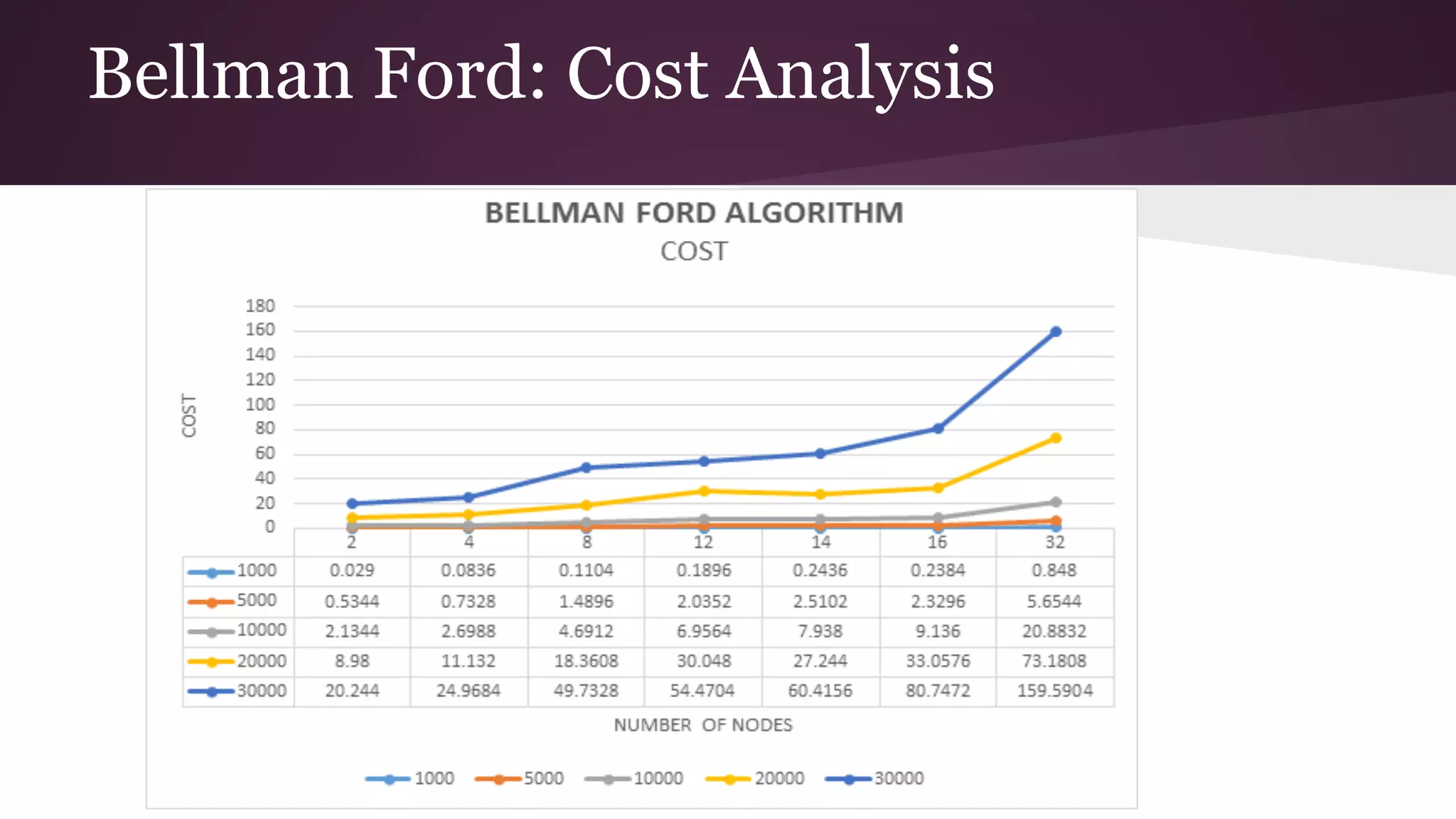
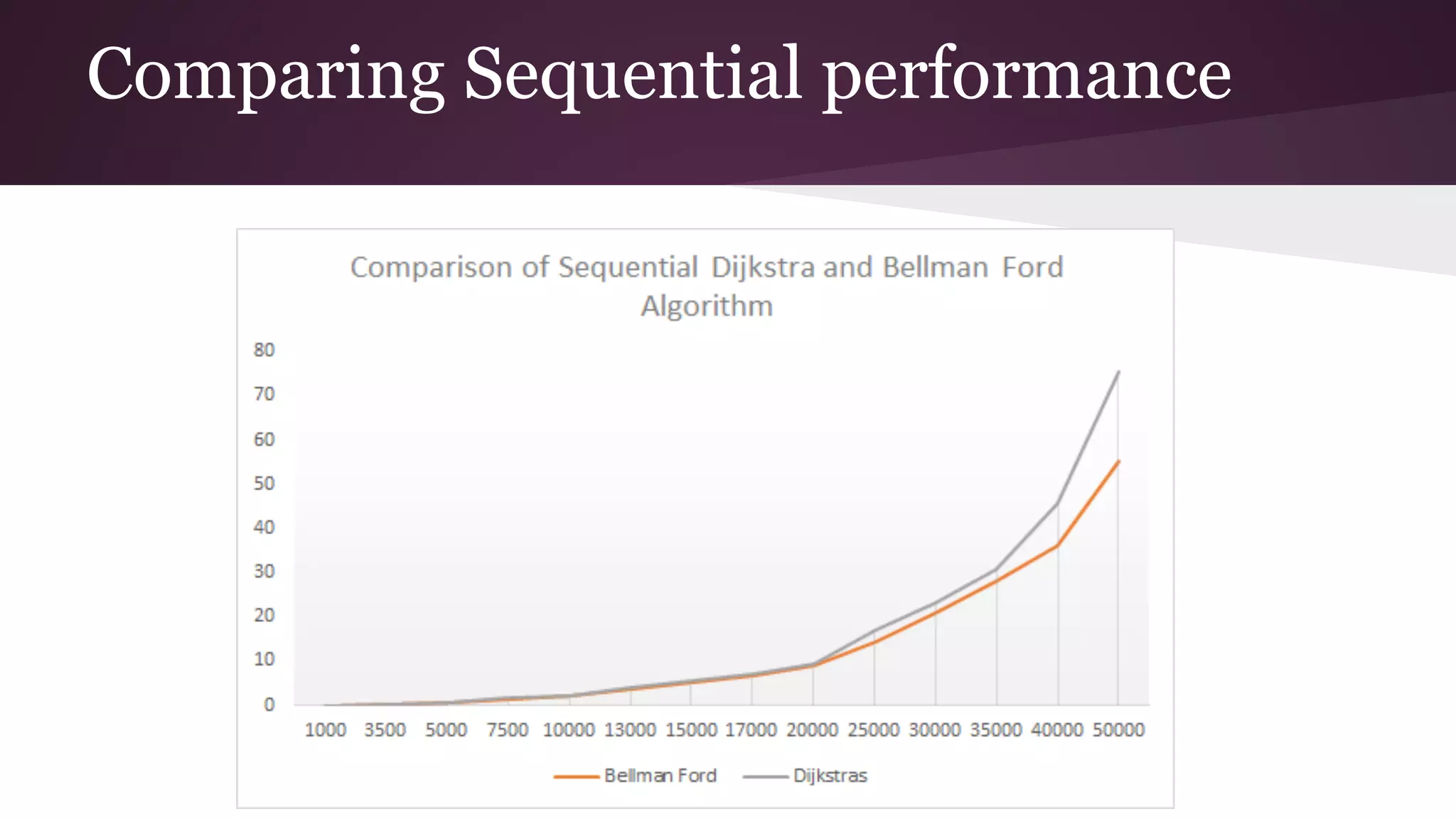
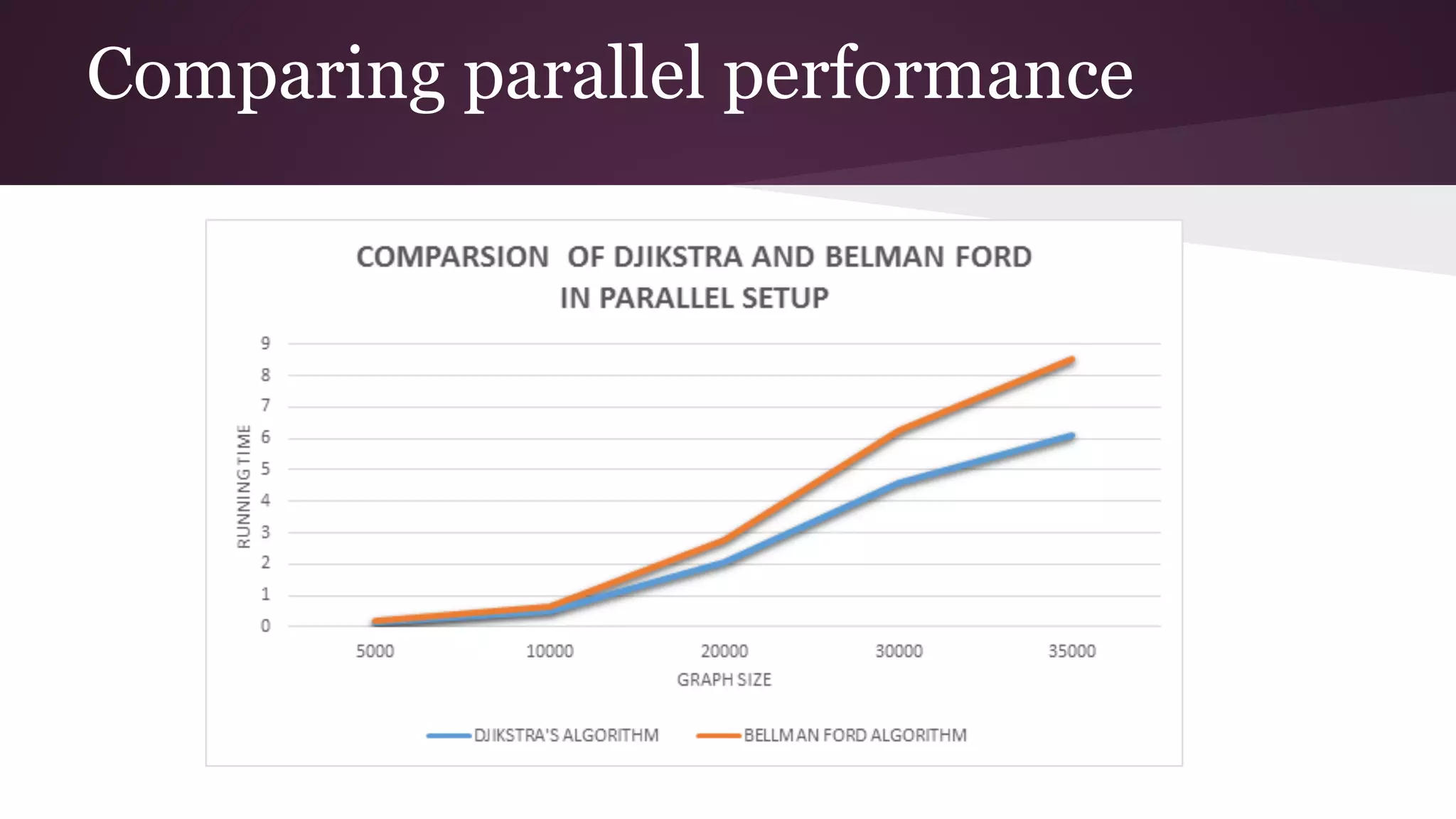
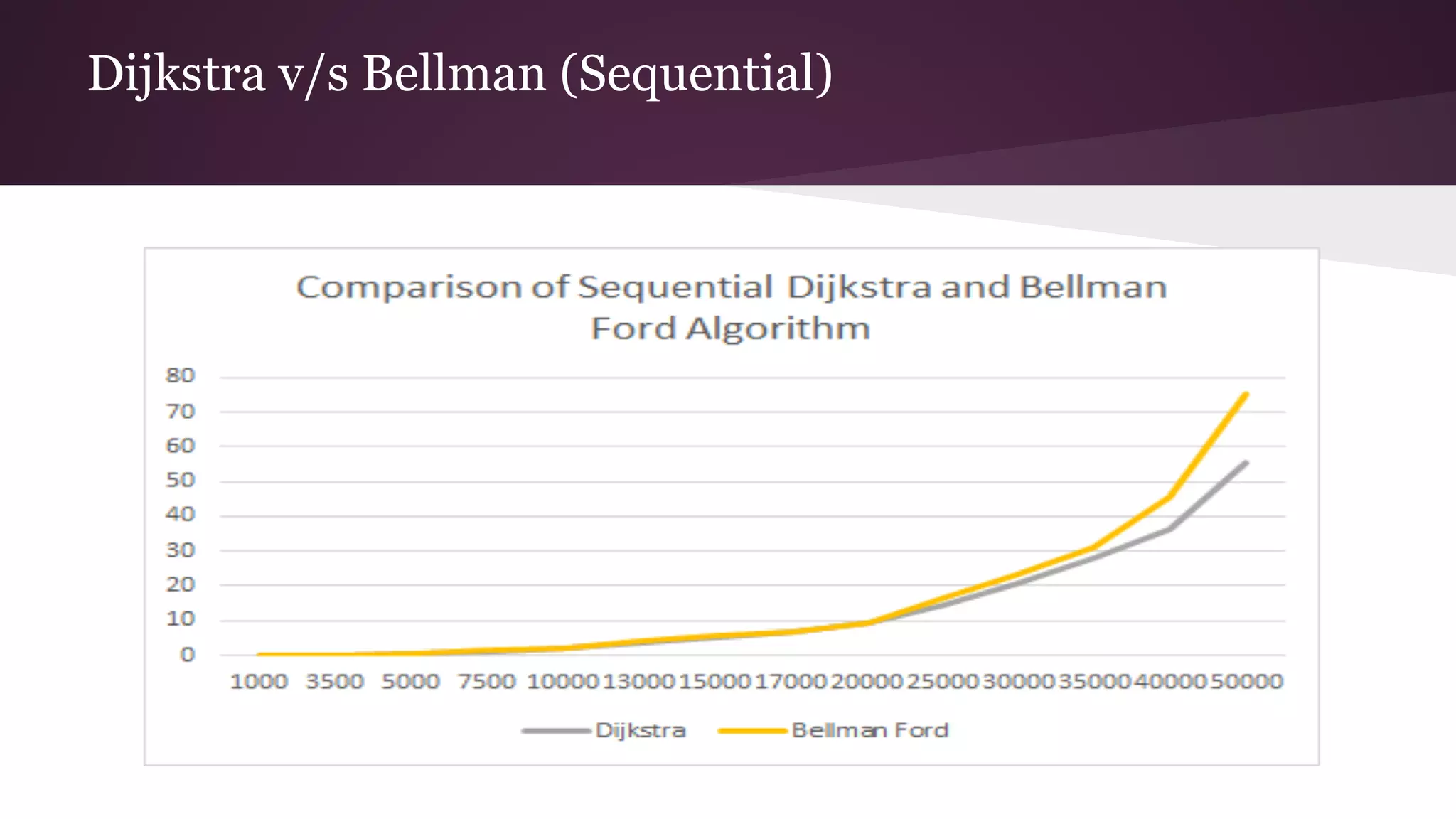
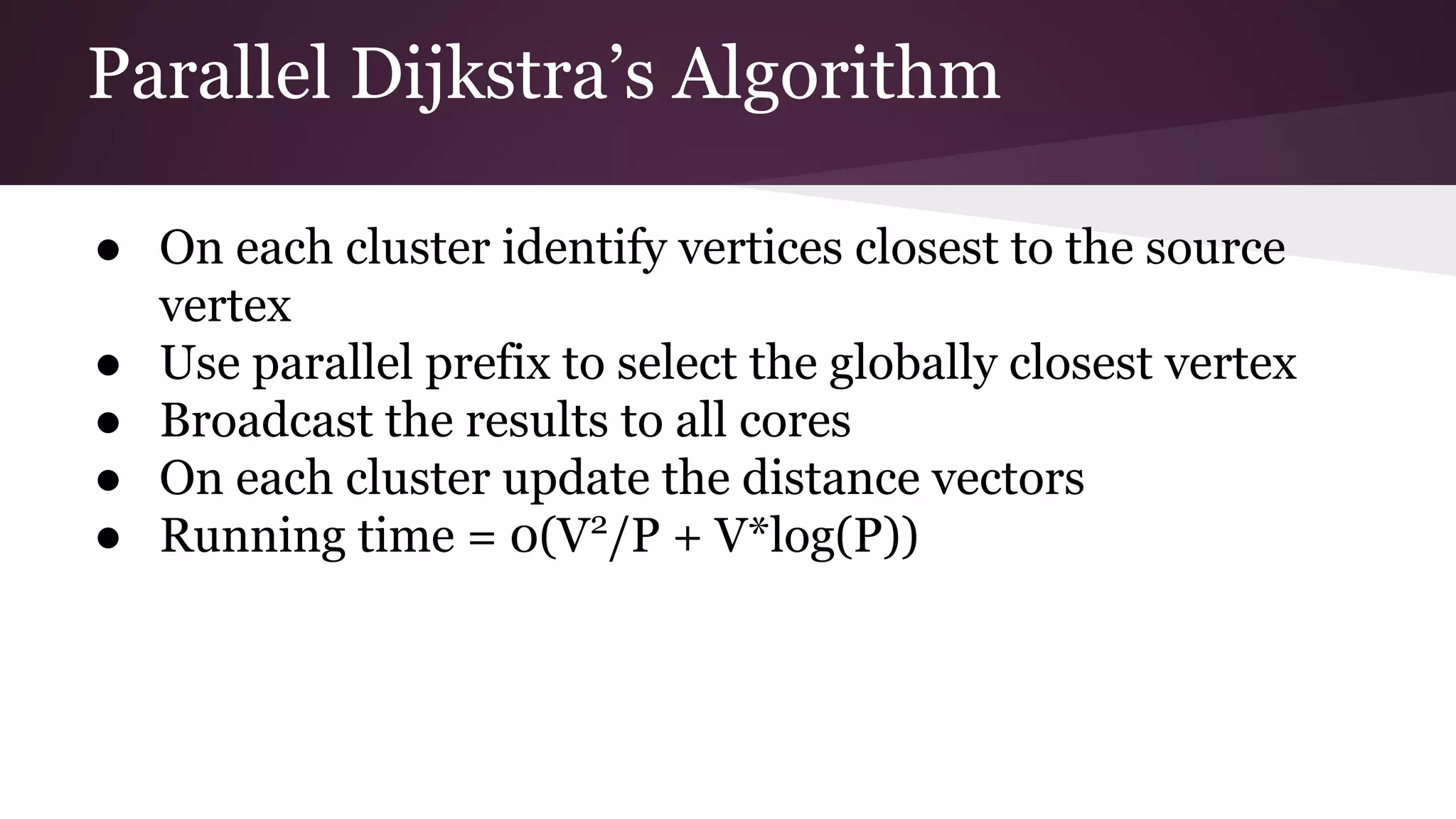
This document compares the parallel implementations of Dijkstra's and Bellman Ford algorithms for solving the single source shortest path problem in graphs. It discusses distributing graph nodes across processors, using parallel prefix to find the shortest path, and broadcasting results. Performance analyses show Dijkstra's and Bellman Ford have near-linear speedups and better scaling to larger graphs in parallel compared to sequential implementations.


![Parallel Approach
1.Allocate
graph nodes to
different processors.
2.Shortest distance is
computed using
parallel prefix.
3. Broadcast the
Result to all nodes after finding
closest path.
❏ MPI_Reduce(MPI_IN_PLACE,nextAN,1,MPI_INT,MPI_SUM,0,MPI_COMM_WORLD);
❏ MPI_Bcast( G->node[0], G->N*G->N, MPI_CHAR, 0, MPI_COMM_WORLD);](https://image.slidesharecdn.com/cse633-140418112400-phpapp01/75/CSE633-10-2048.jpg)