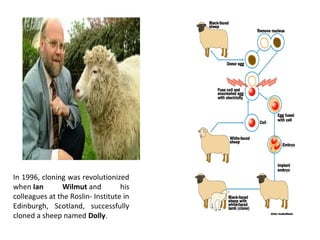
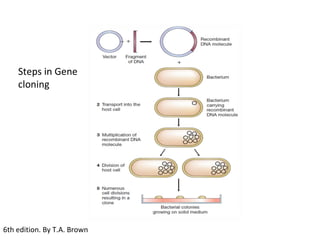
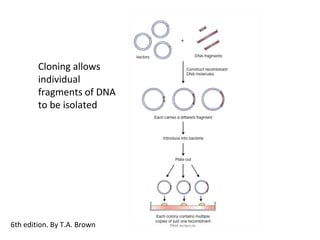
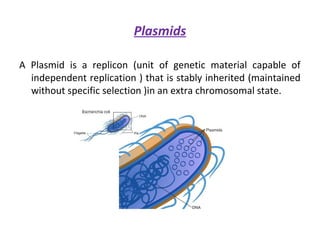
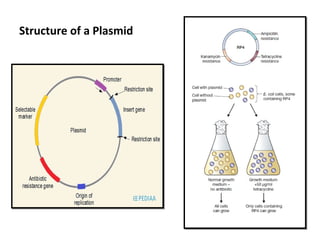
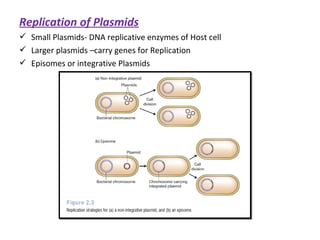
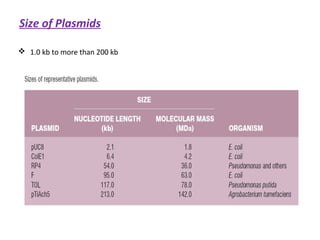
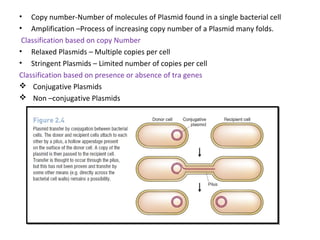
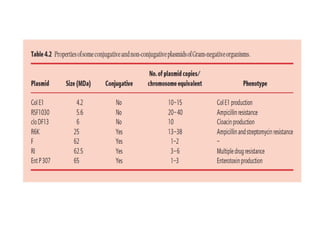
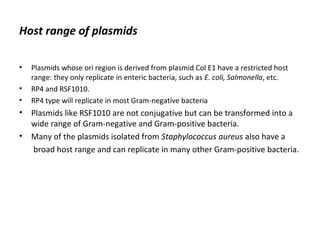
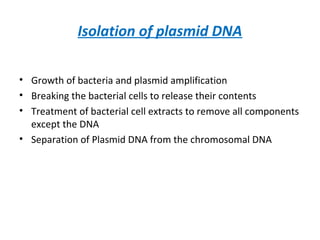
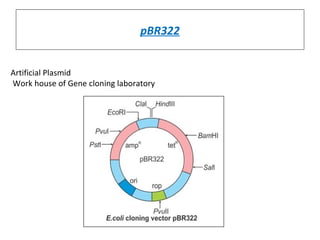
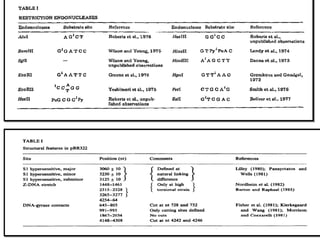
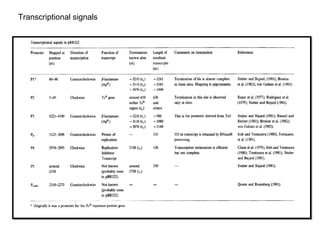
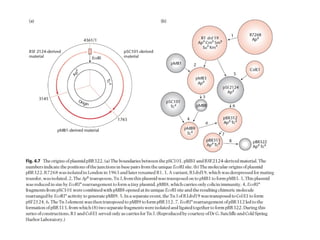
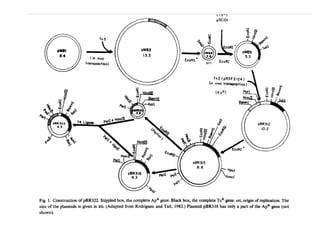
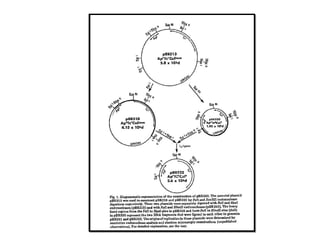
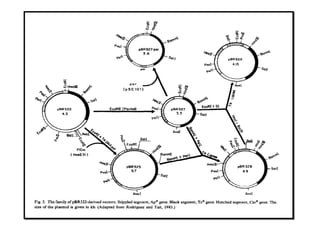
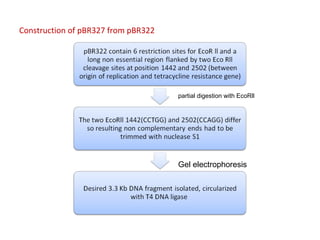
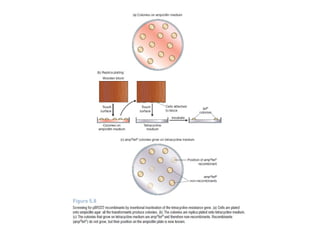
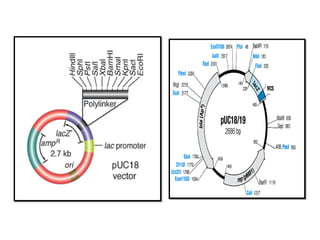
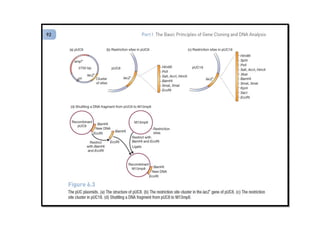
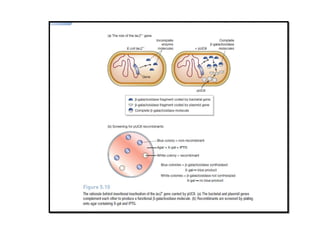
This document discusses gene cloning and various cloning vectors. It provides details about plasmid vectors such as pBR322 and pUC18/19. pBR322 was one of the first widely used E. coli cloning vectors, containing genes for ampicillin and tetracycline resistance. It served as a model for prokaryotic transcription and replication studies. The document discusses the structure, properties, replication and uses of pBR322. It also describes the expression vector pUC18/19, which is derived from pBR322 and contains a multiple cloning site for inserting genes between the beta-lactamase gene and a fragment of the lacZ gene.