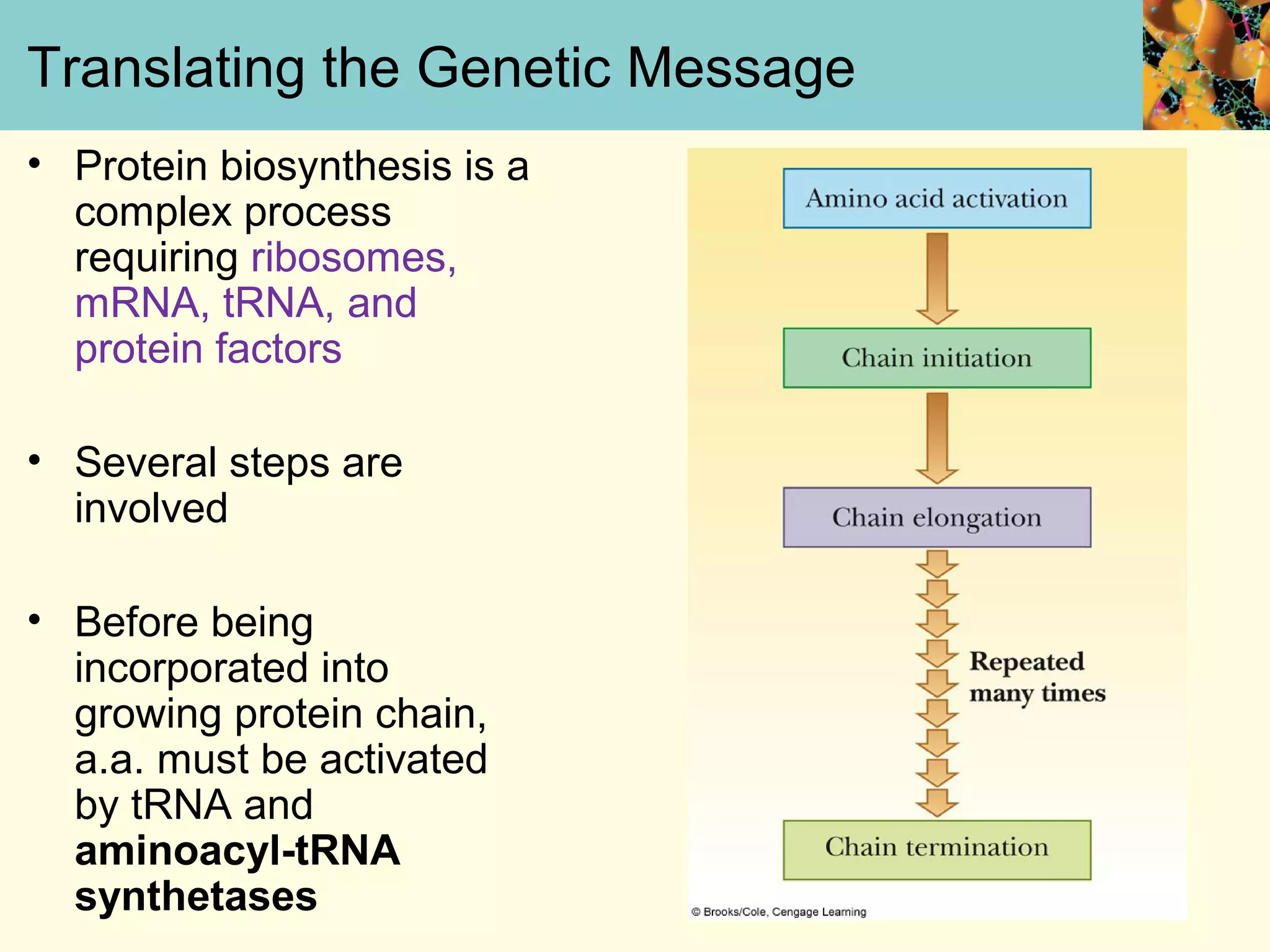
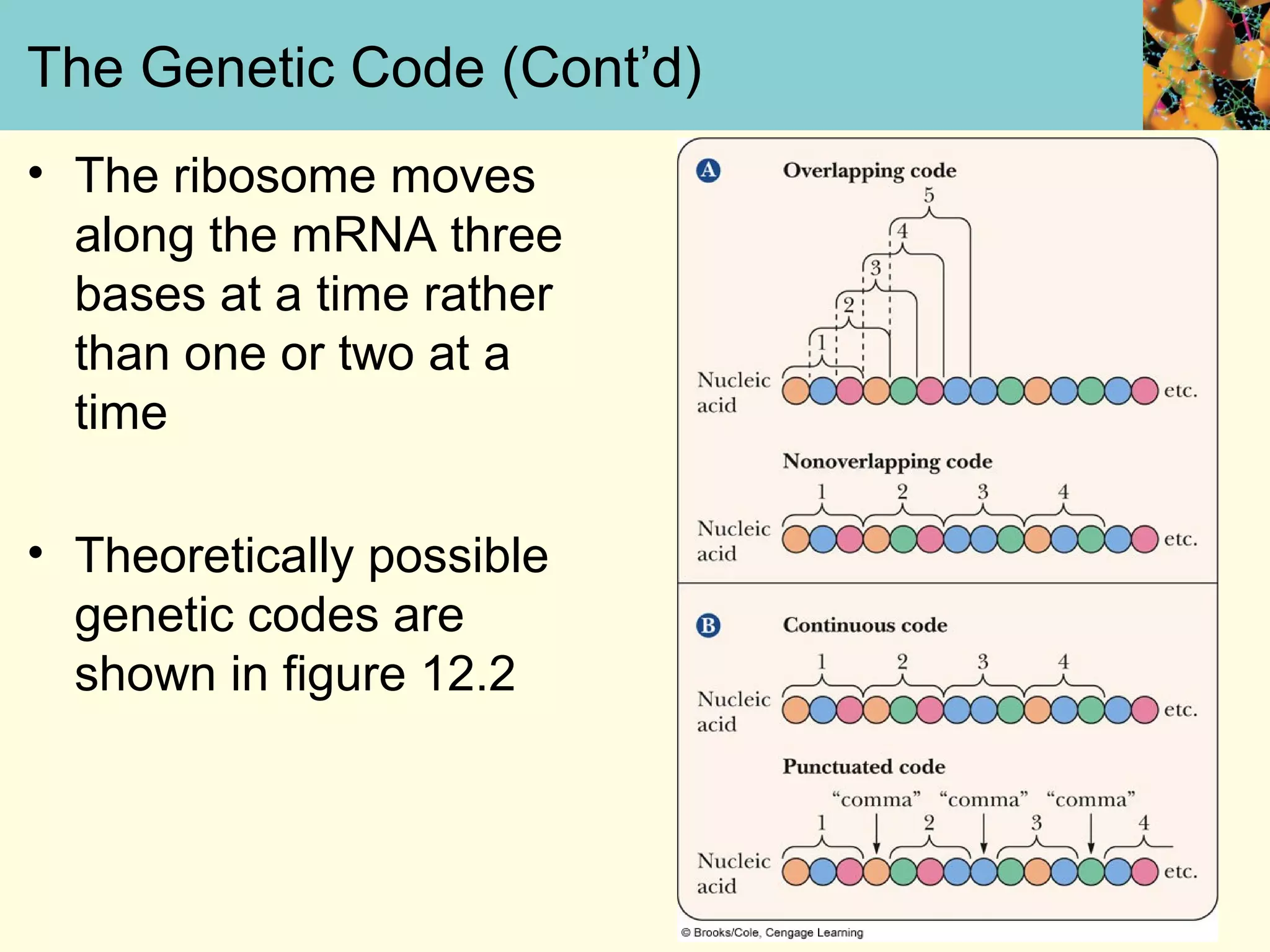
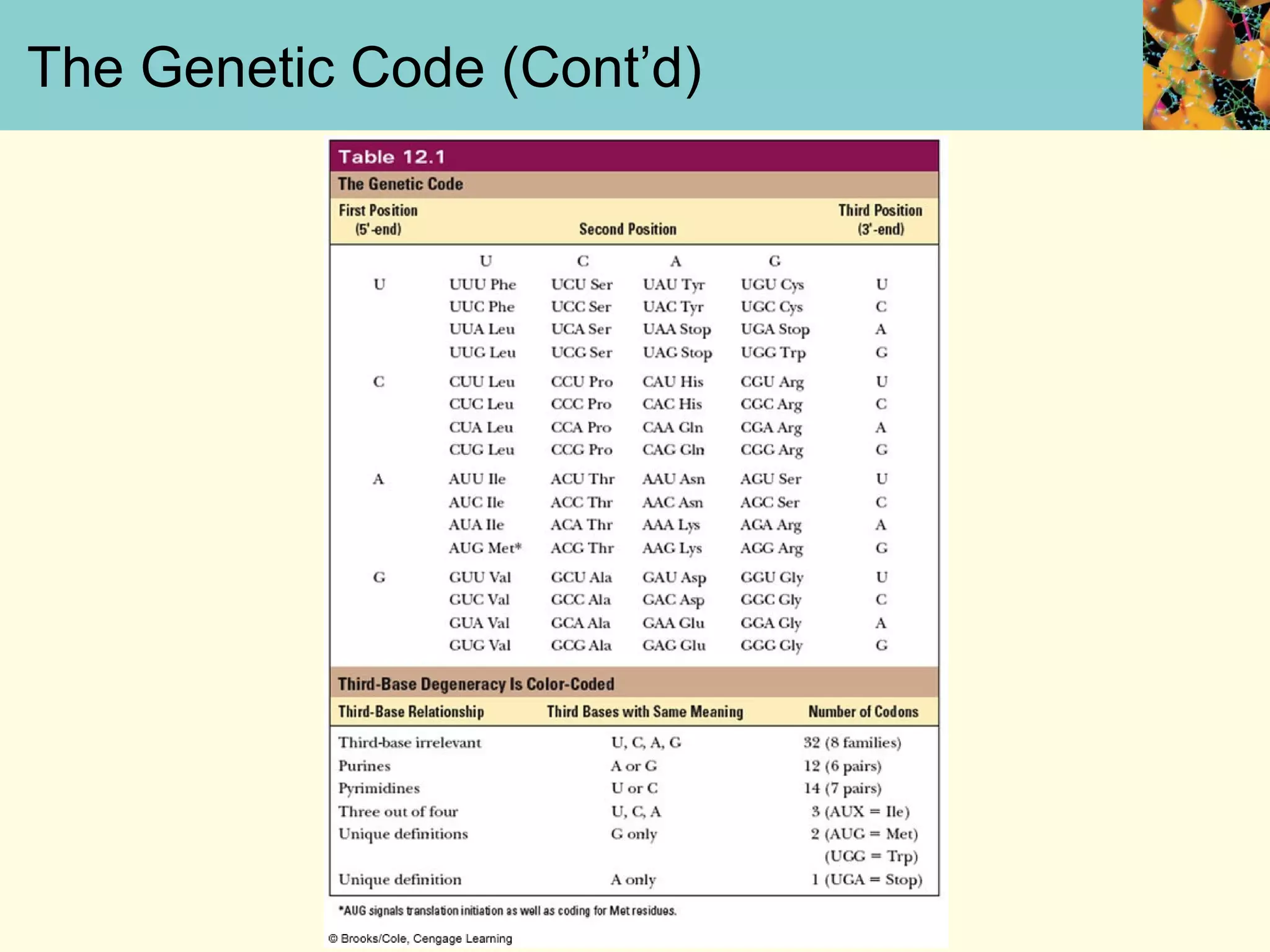
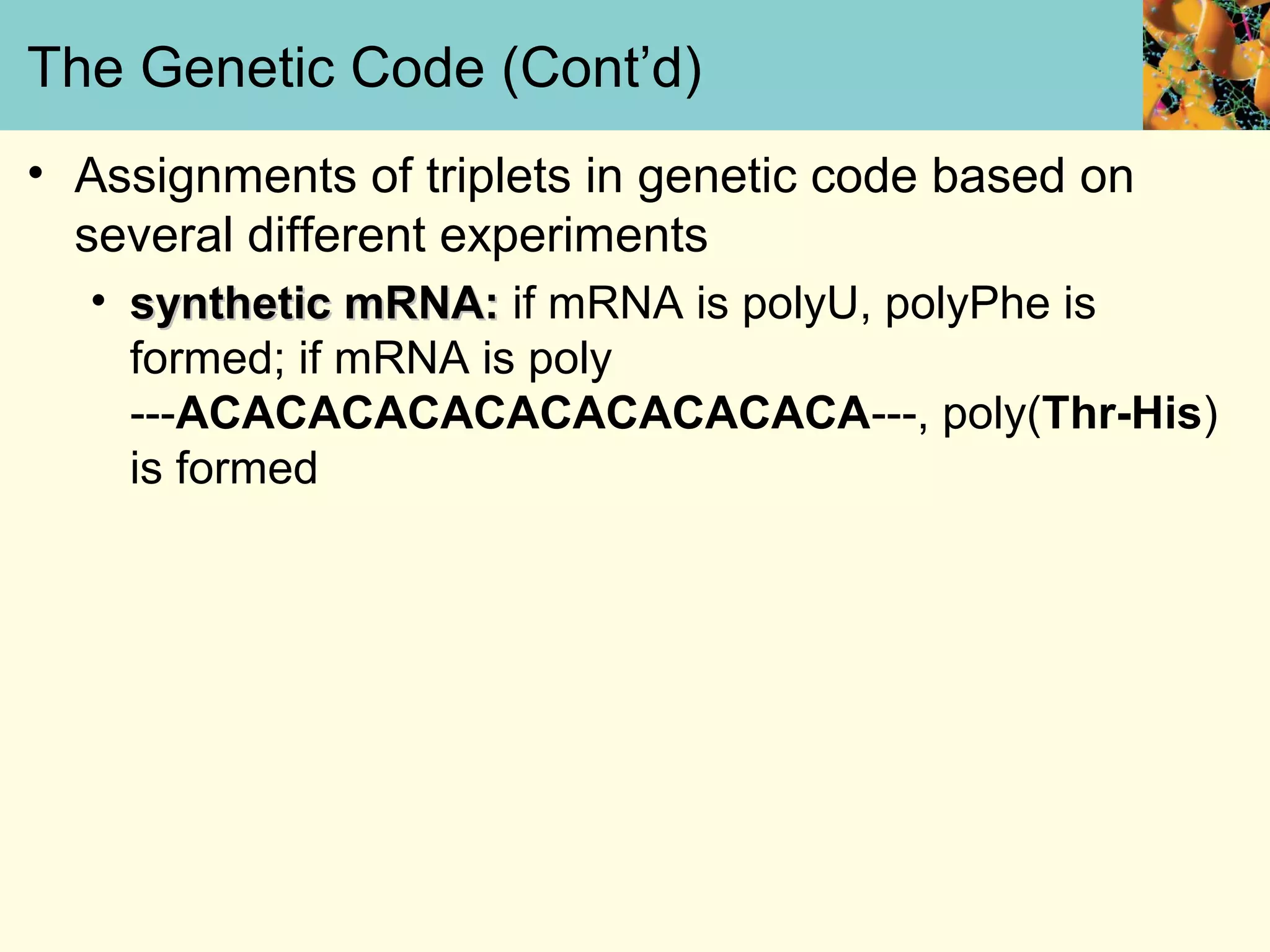
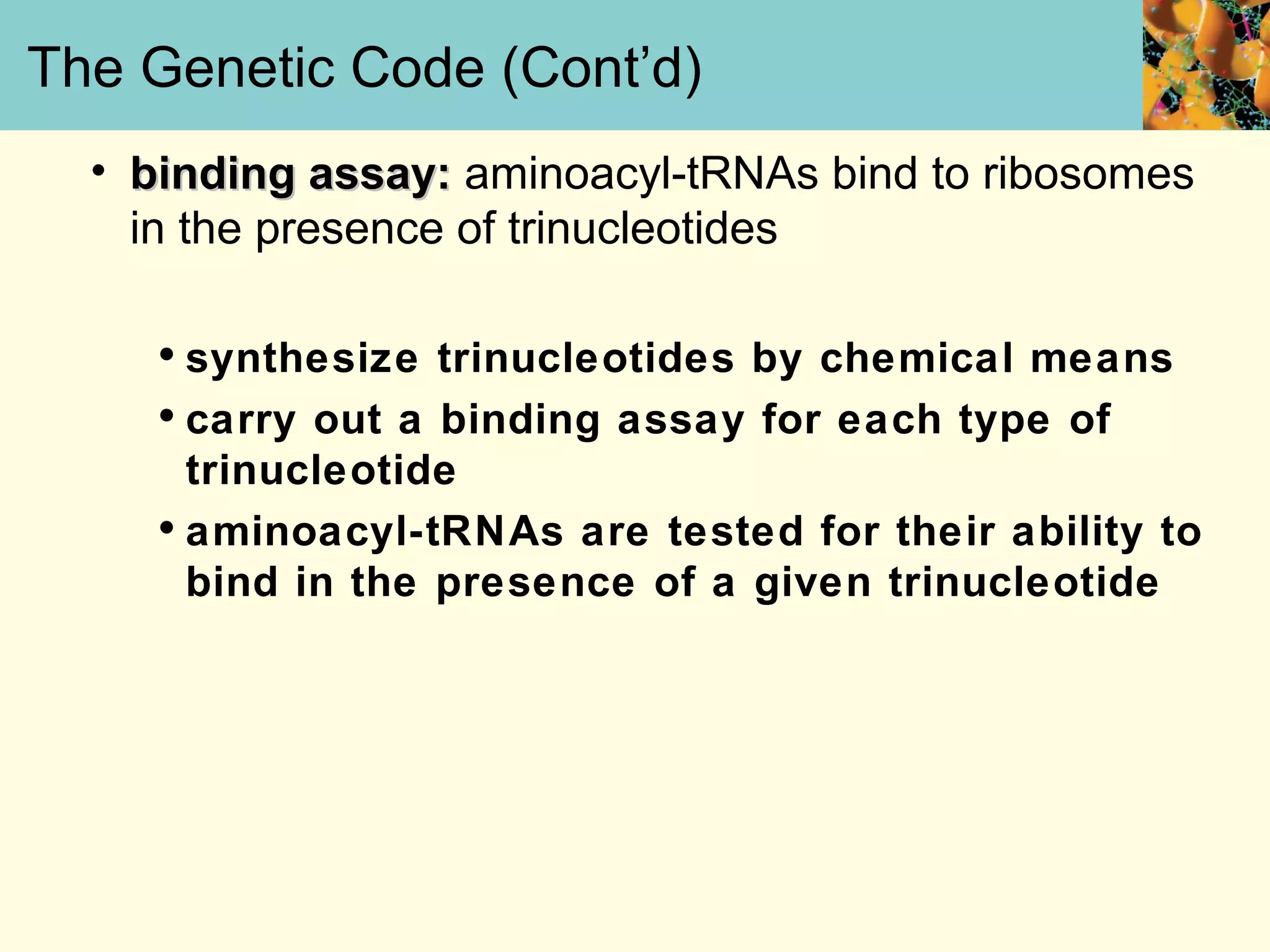
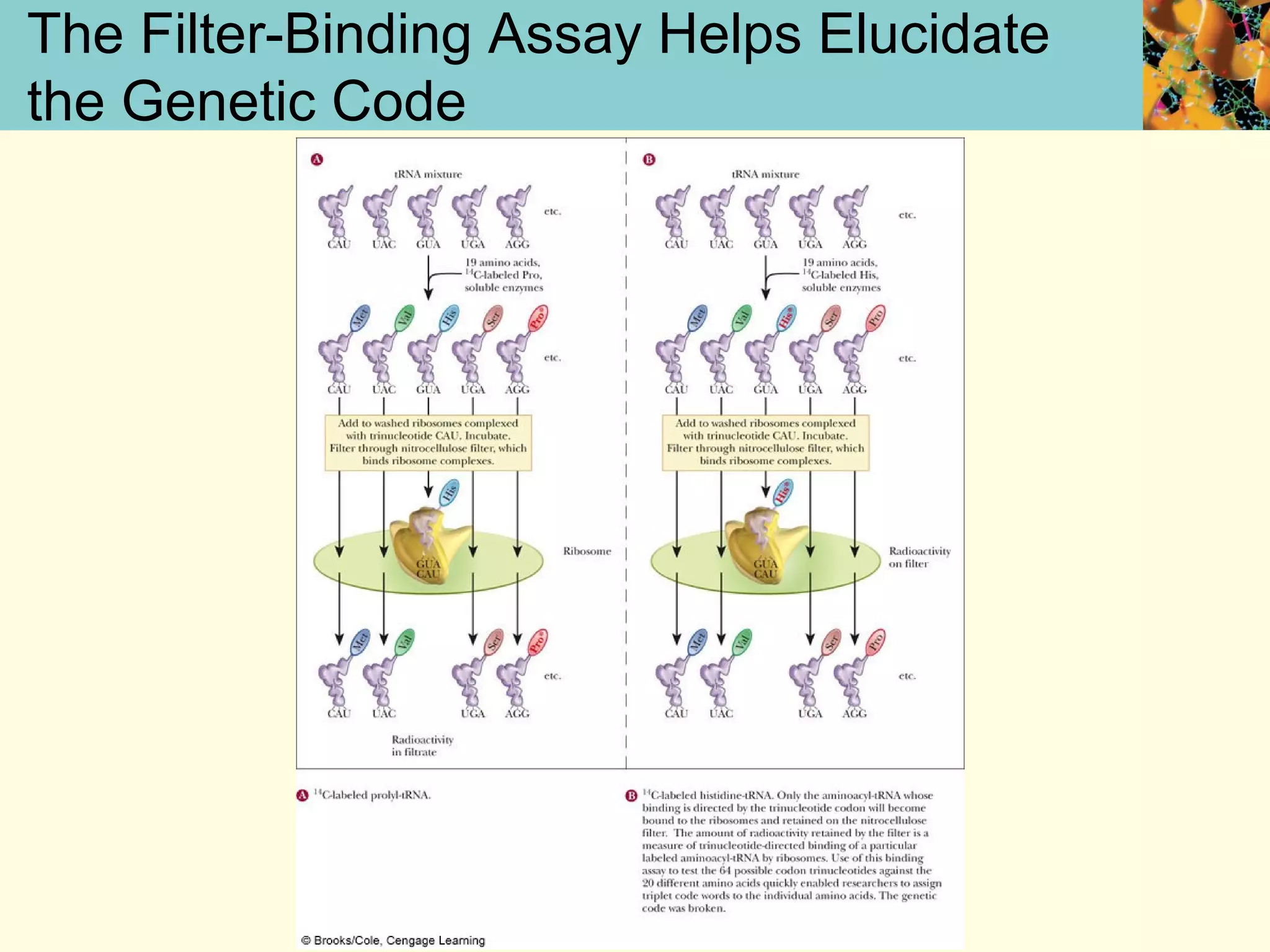
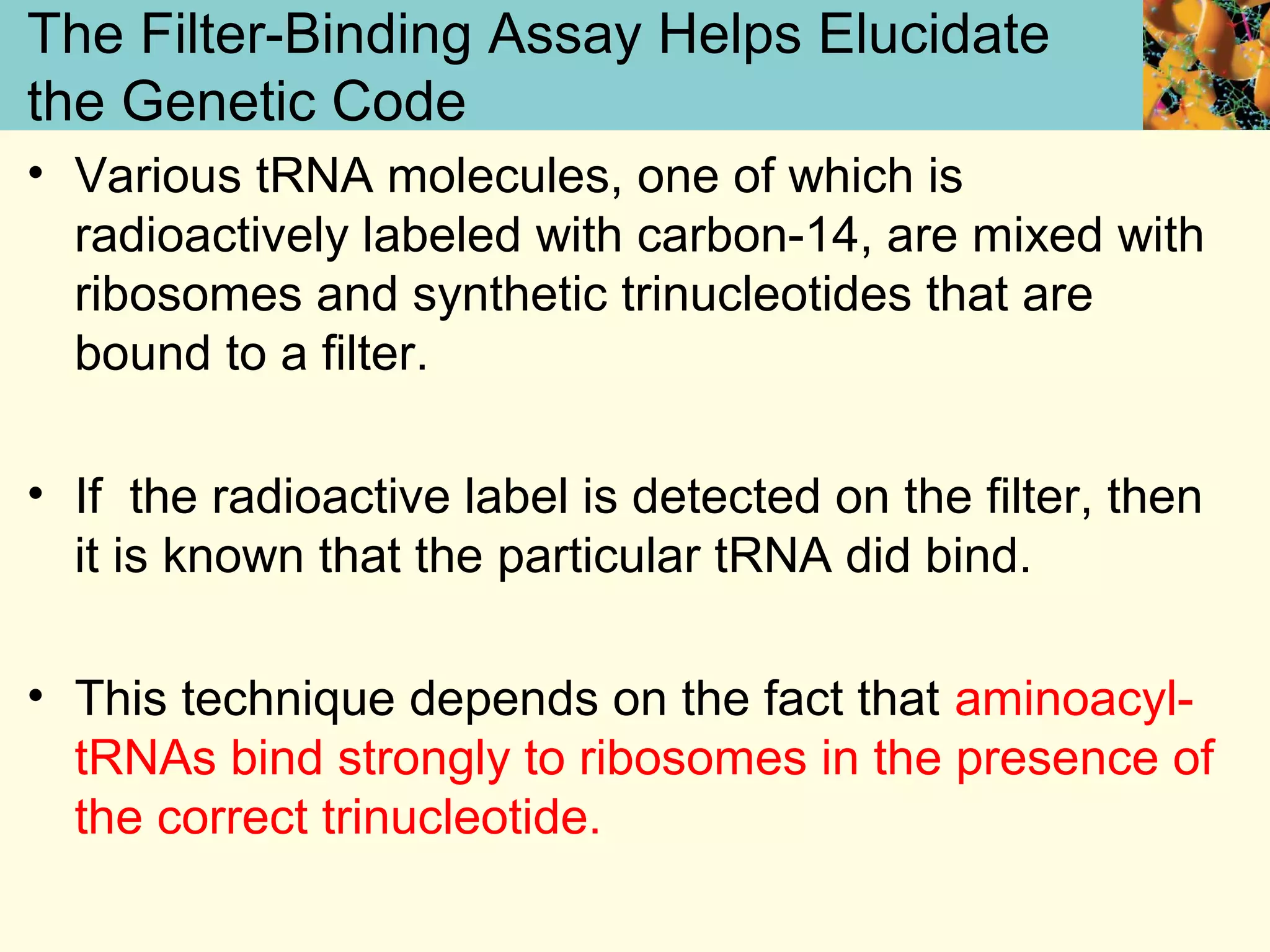
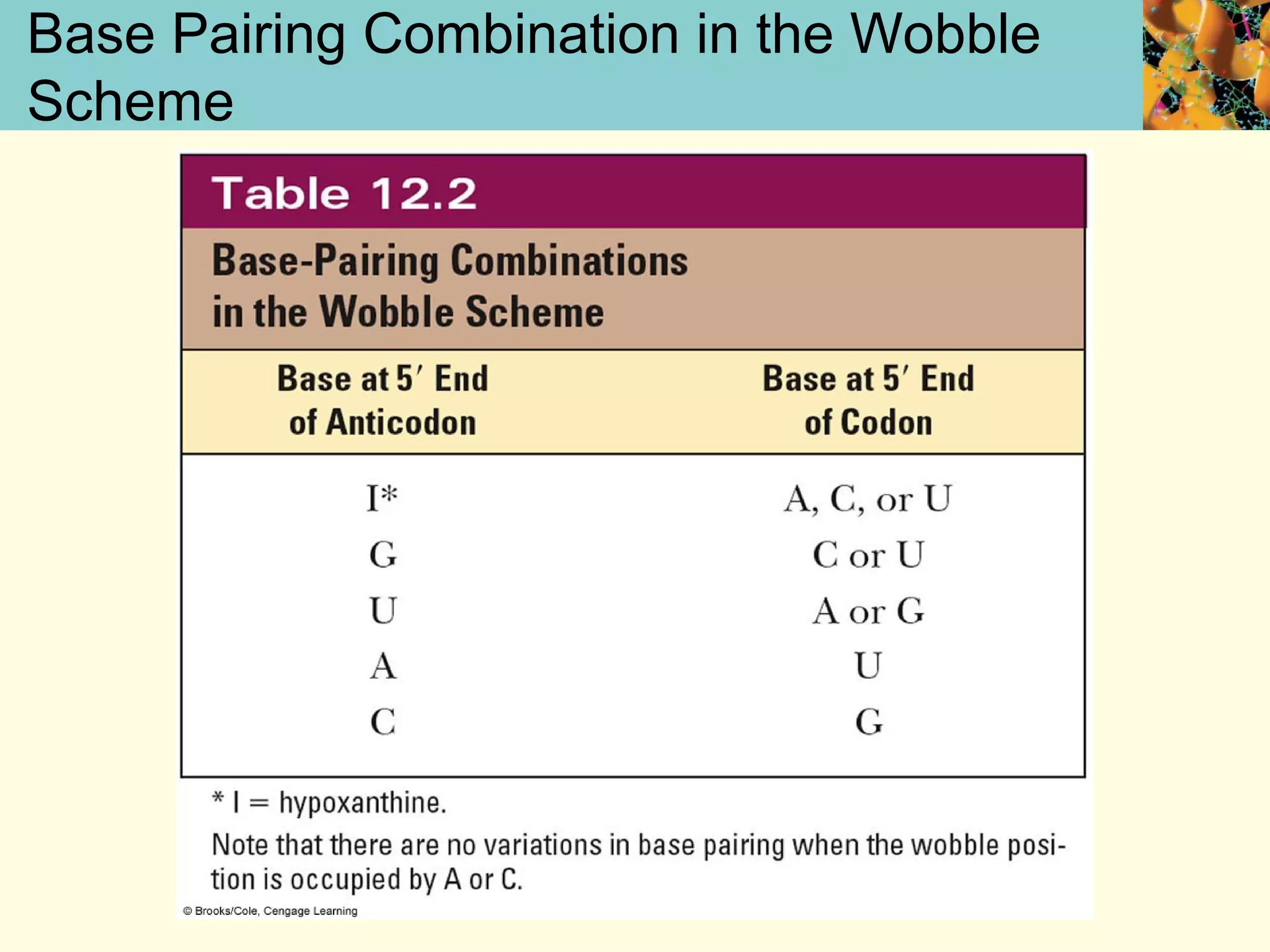
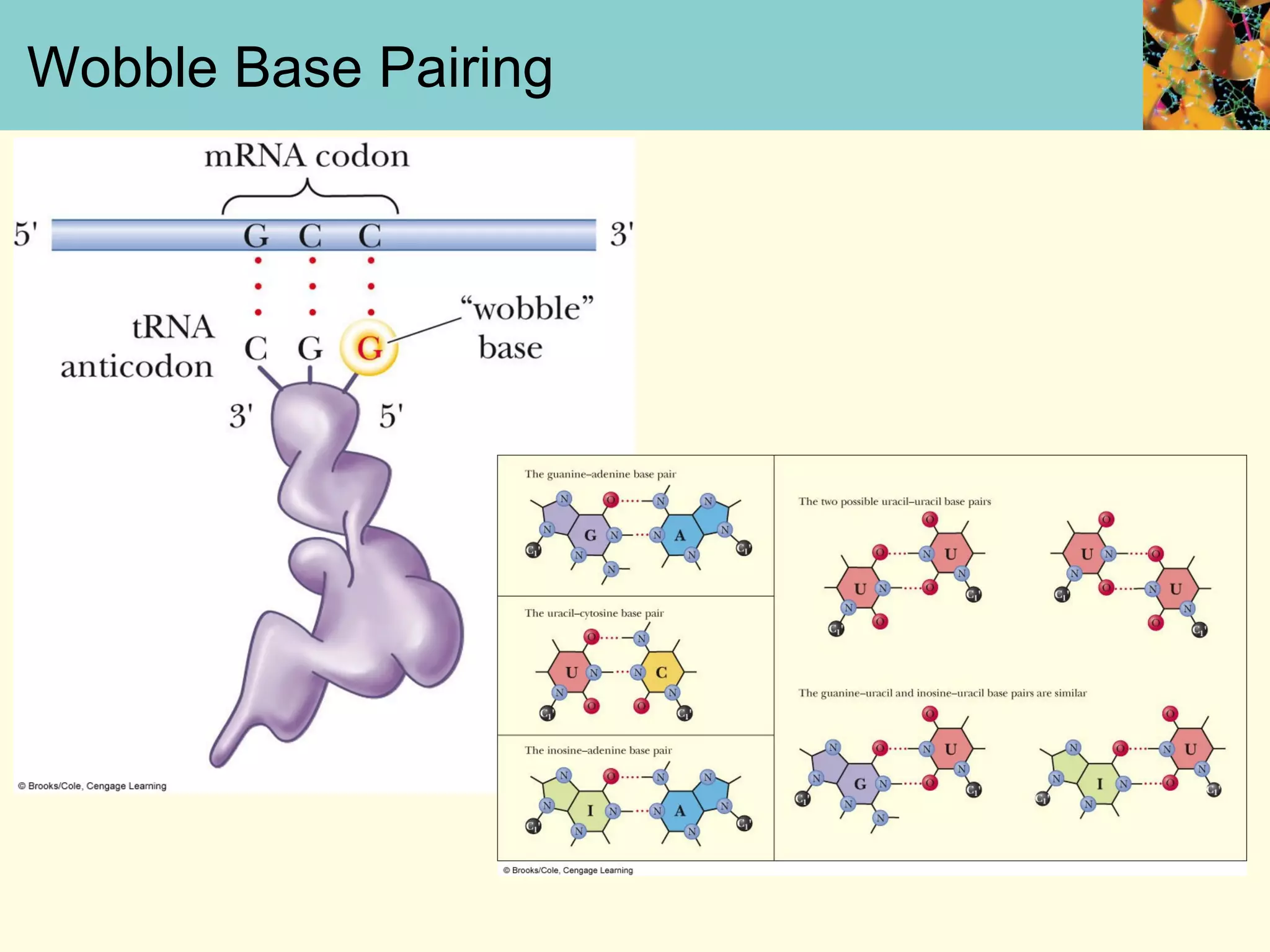
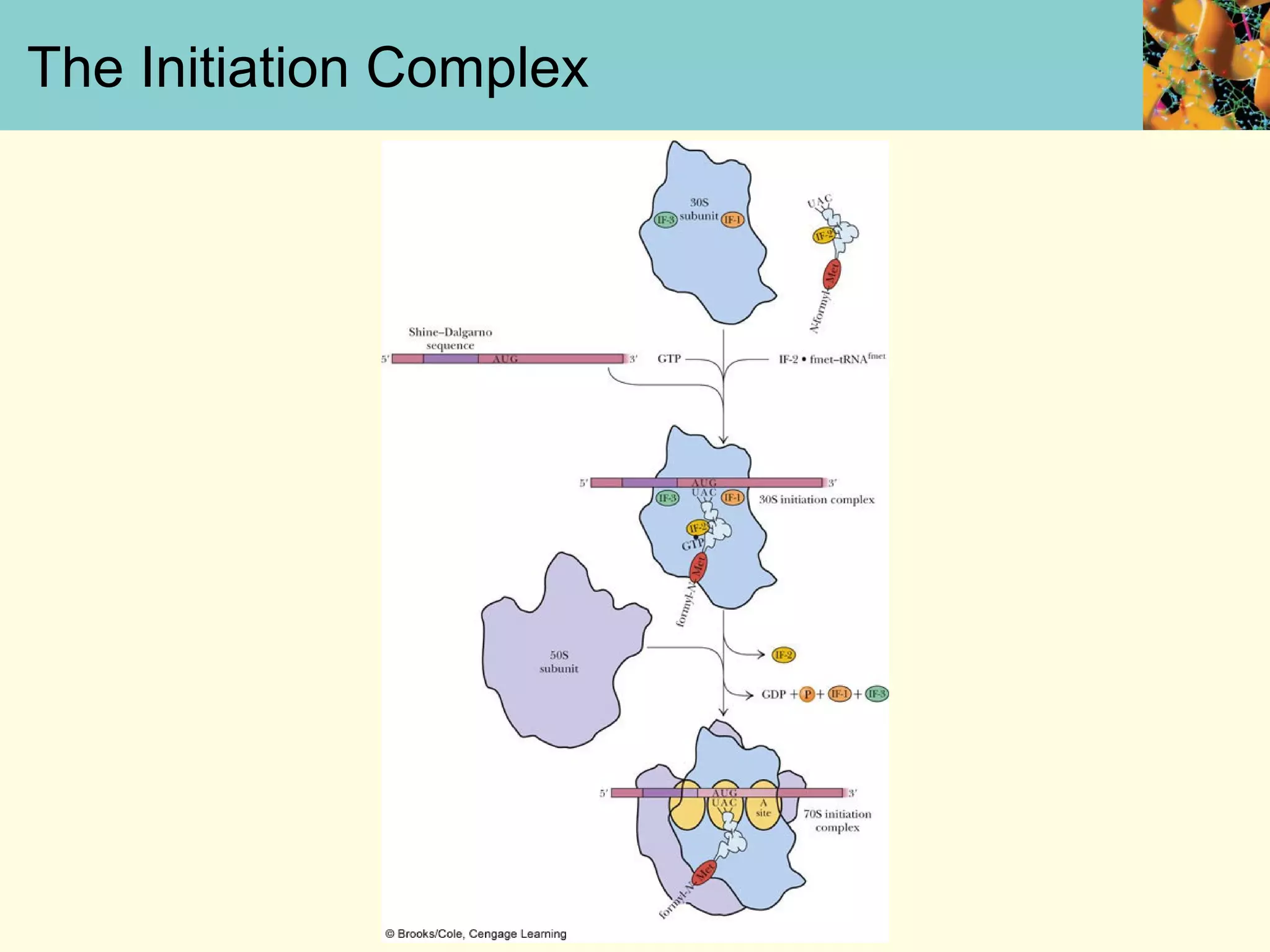
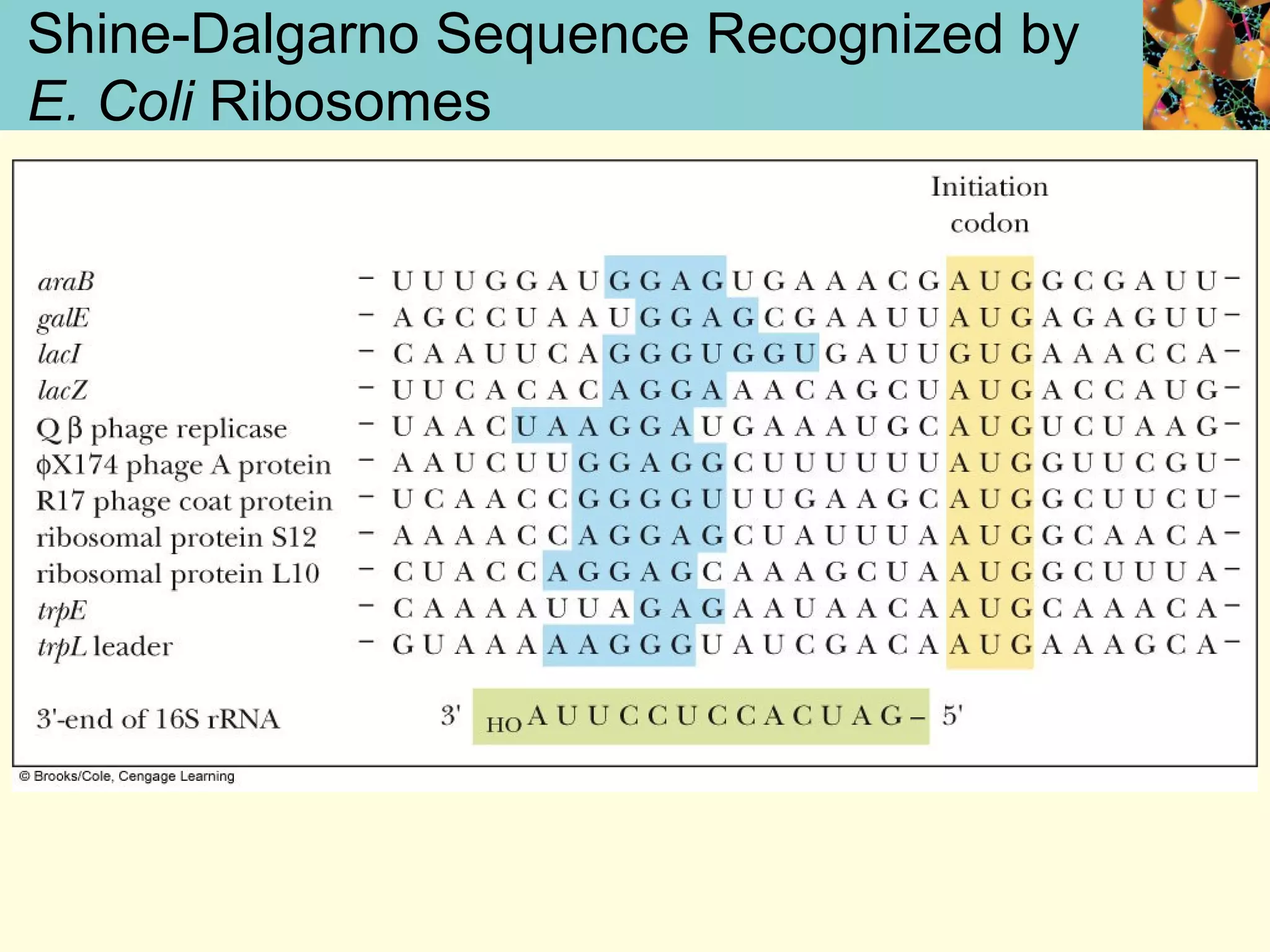
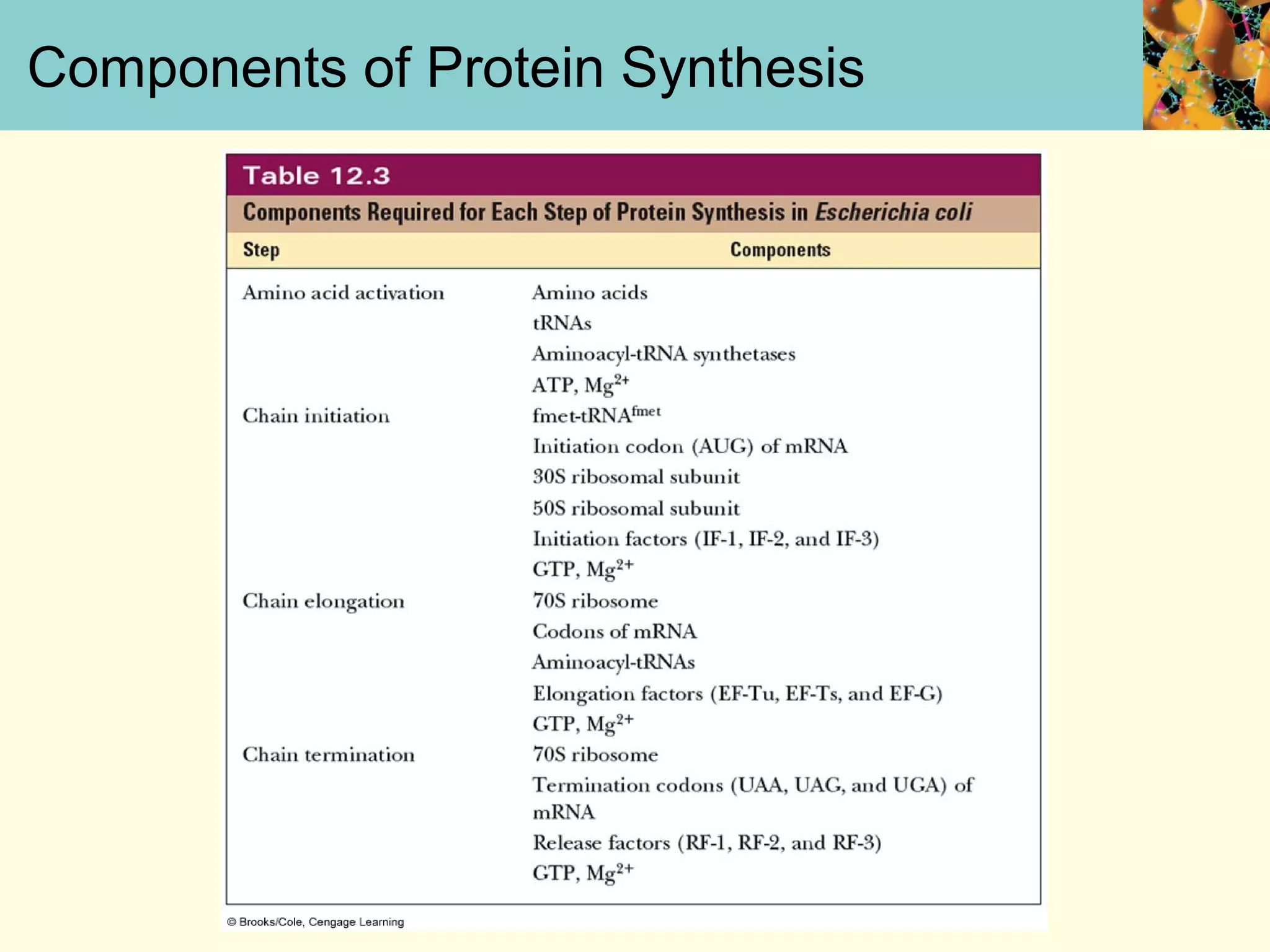
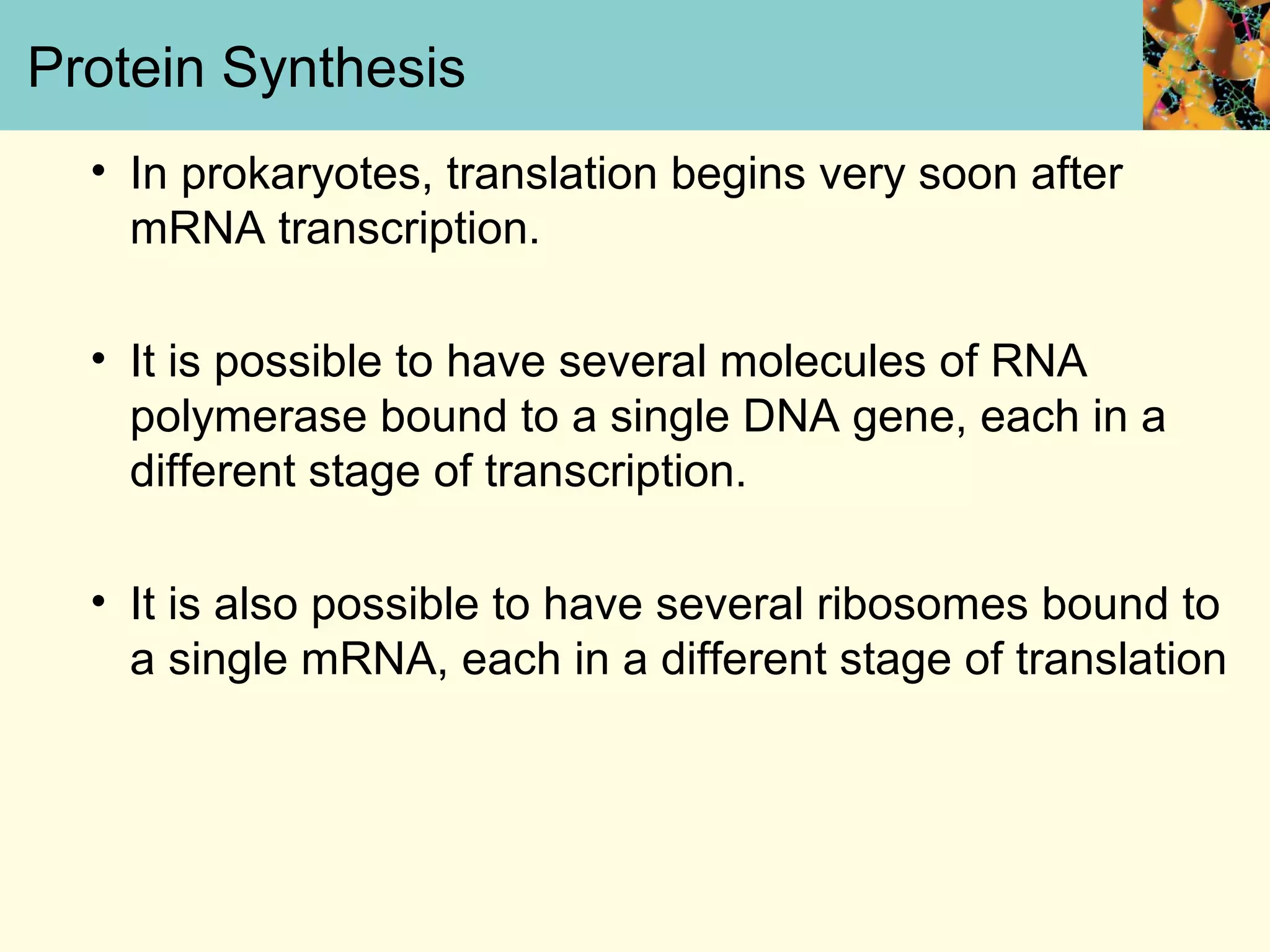
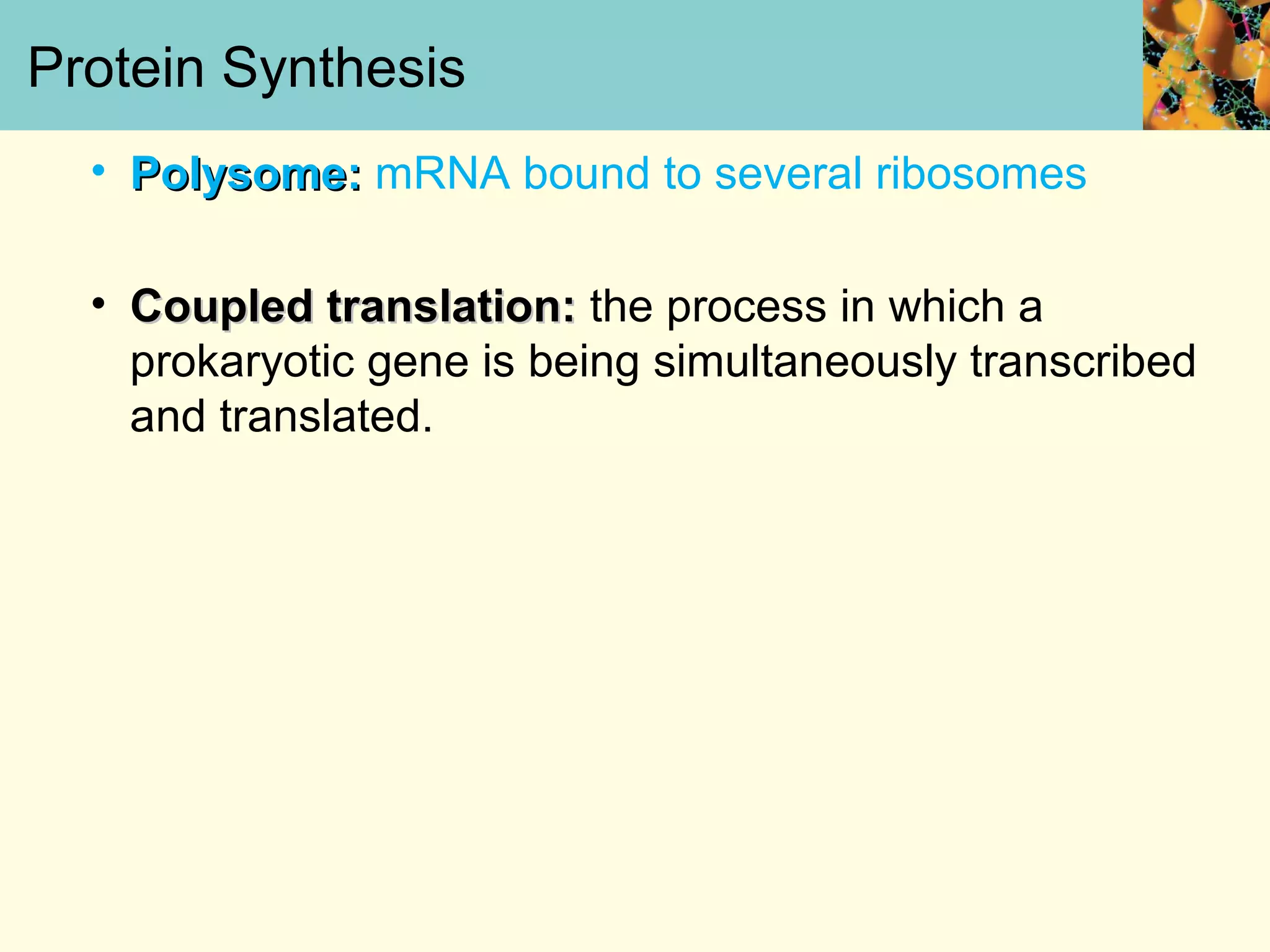
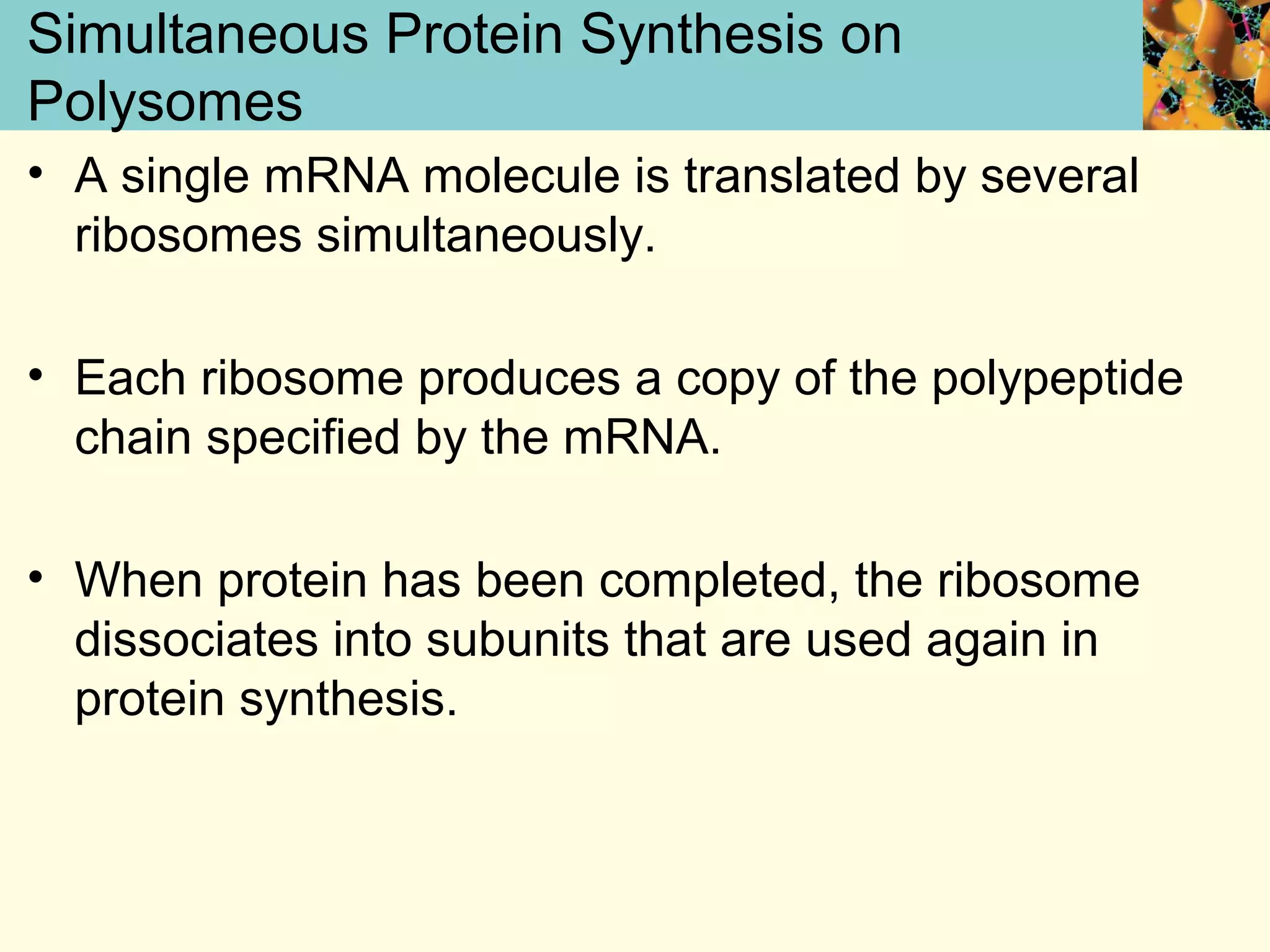
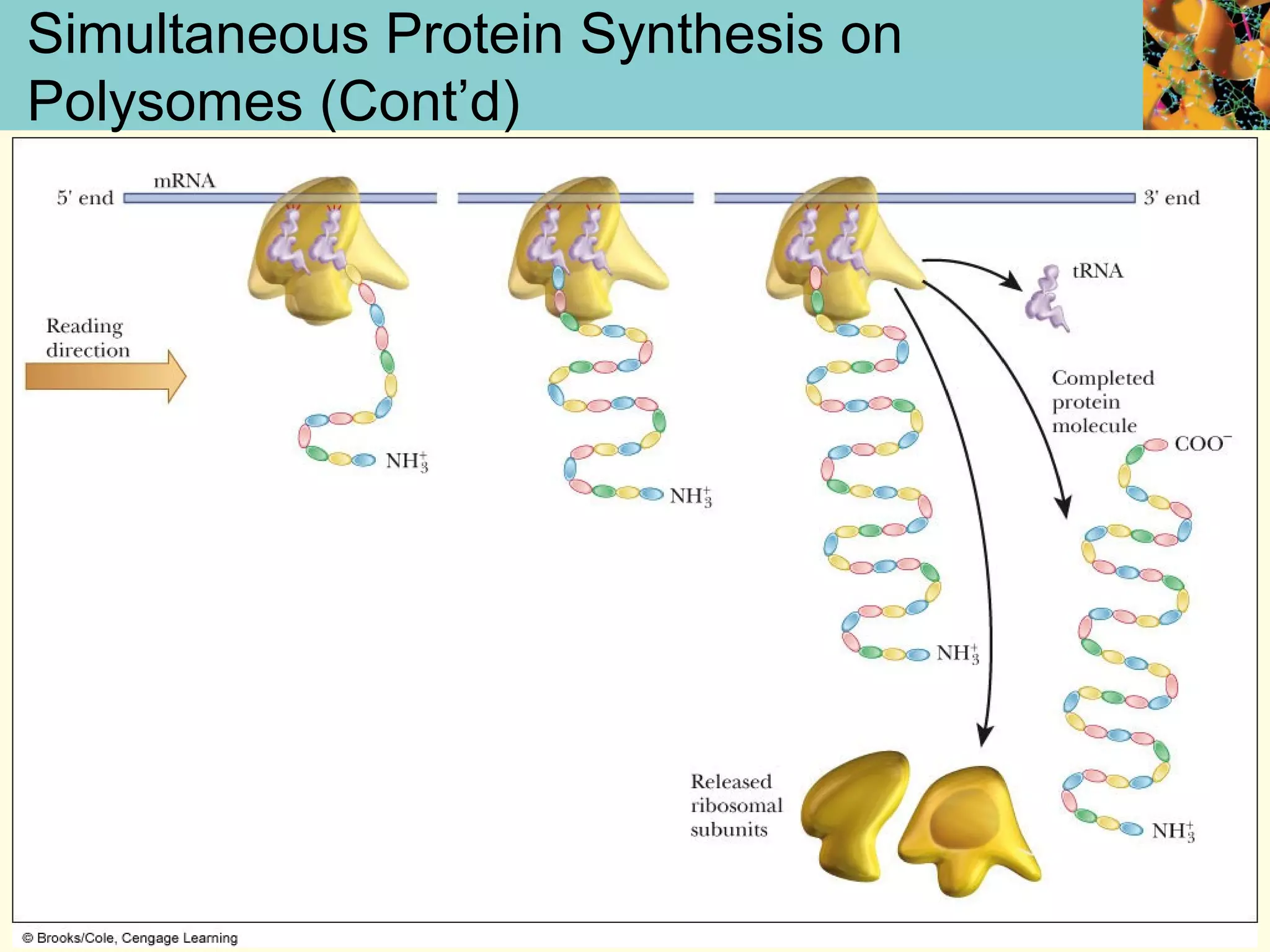
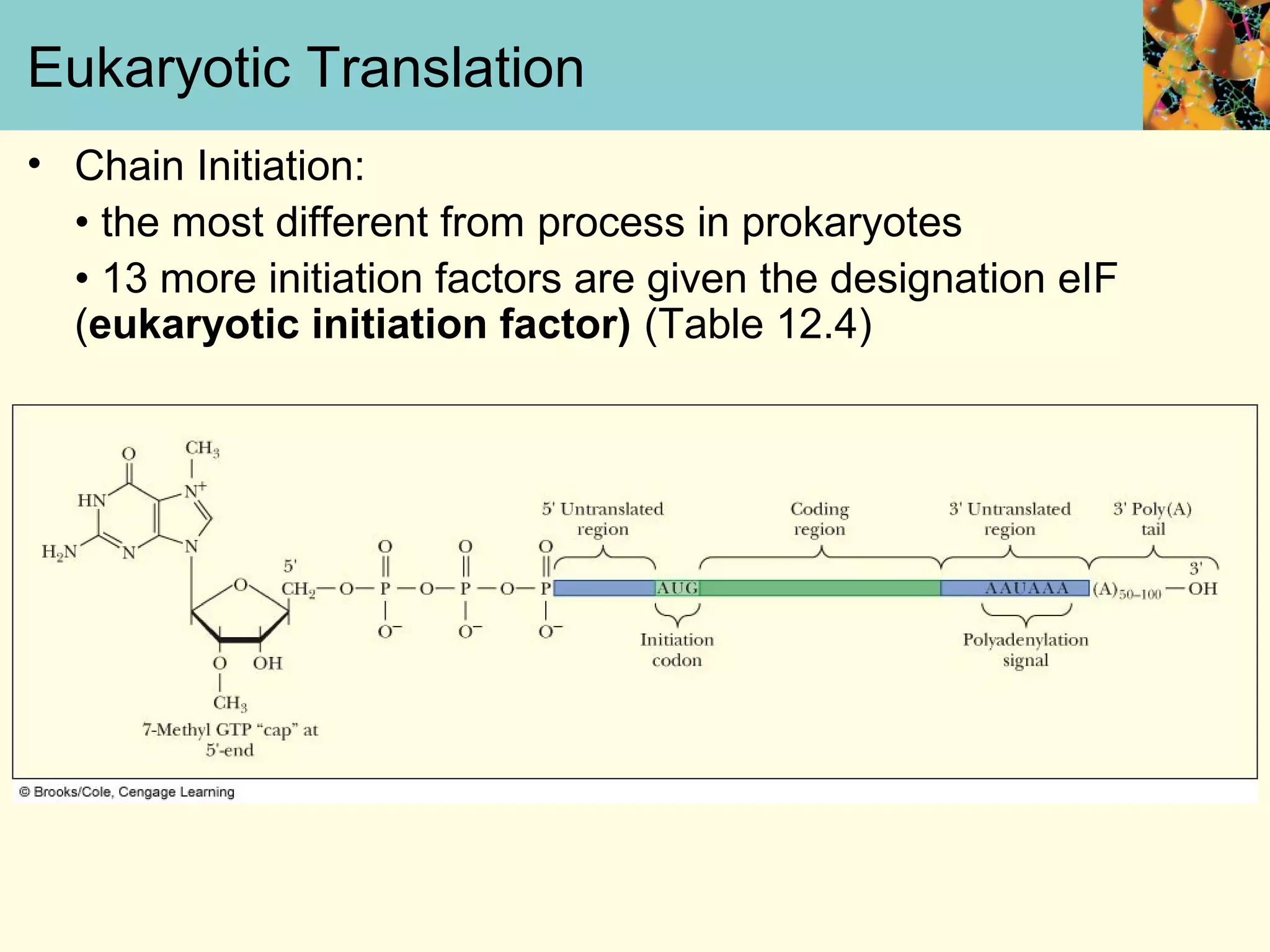
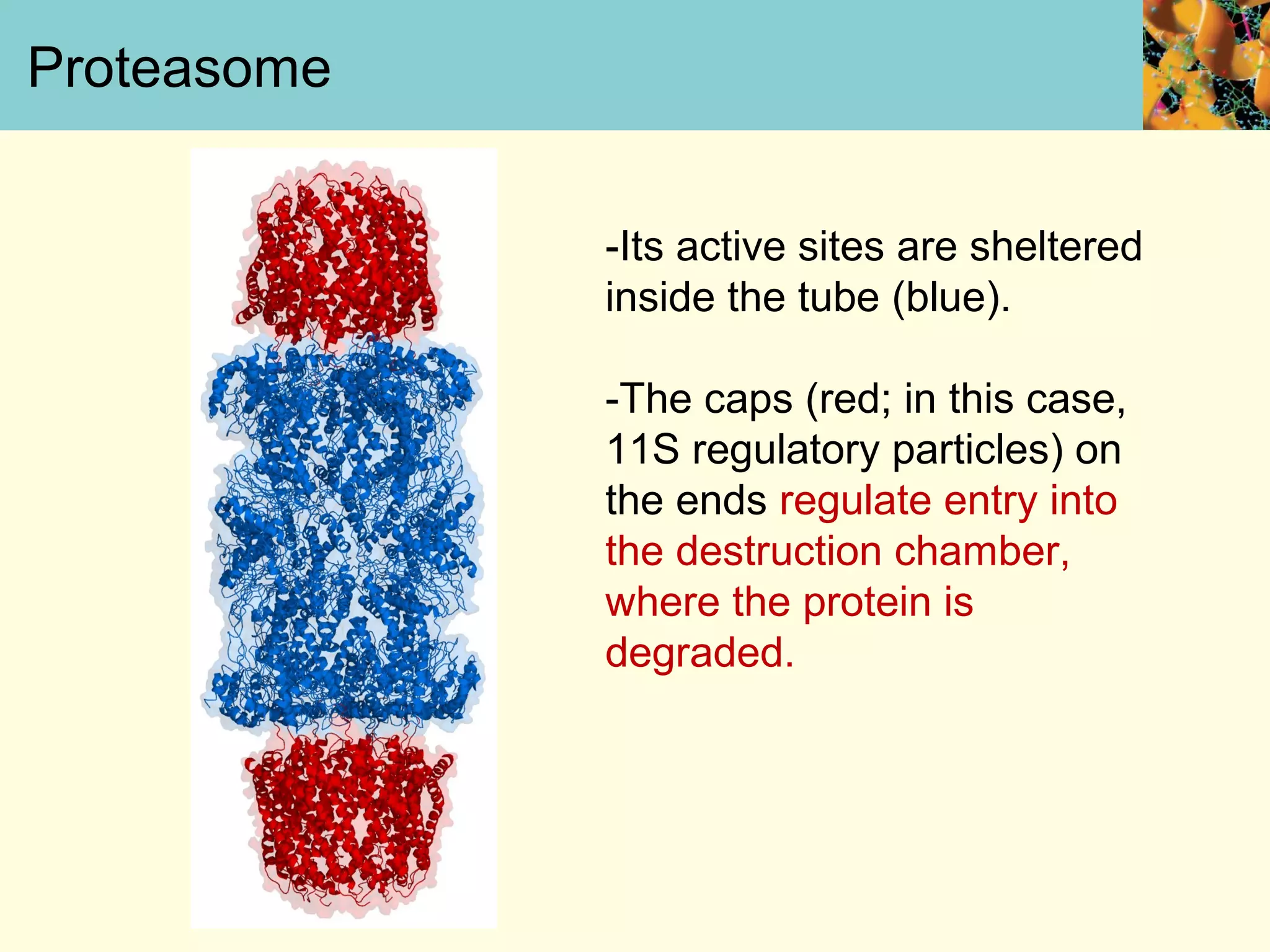
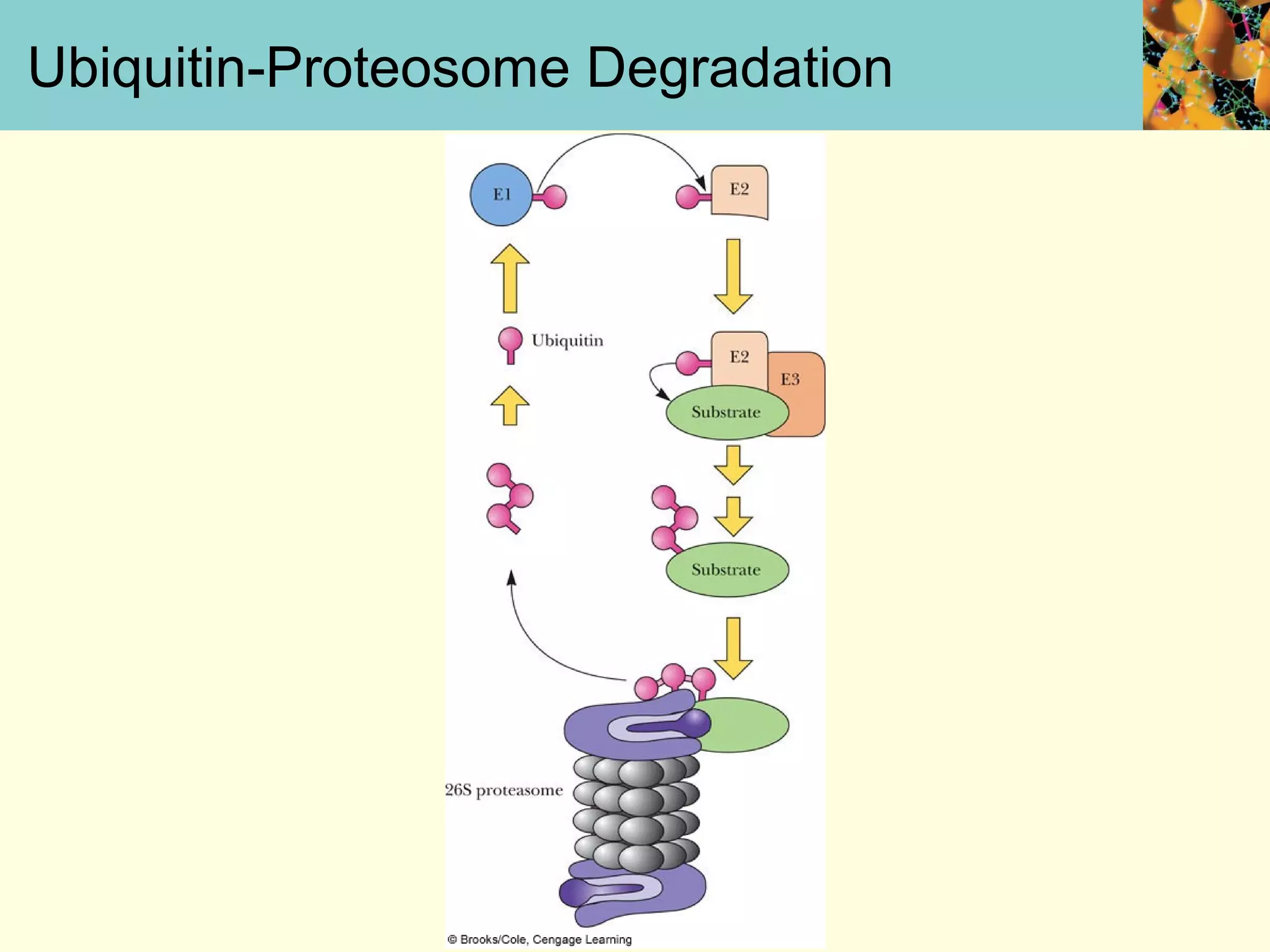
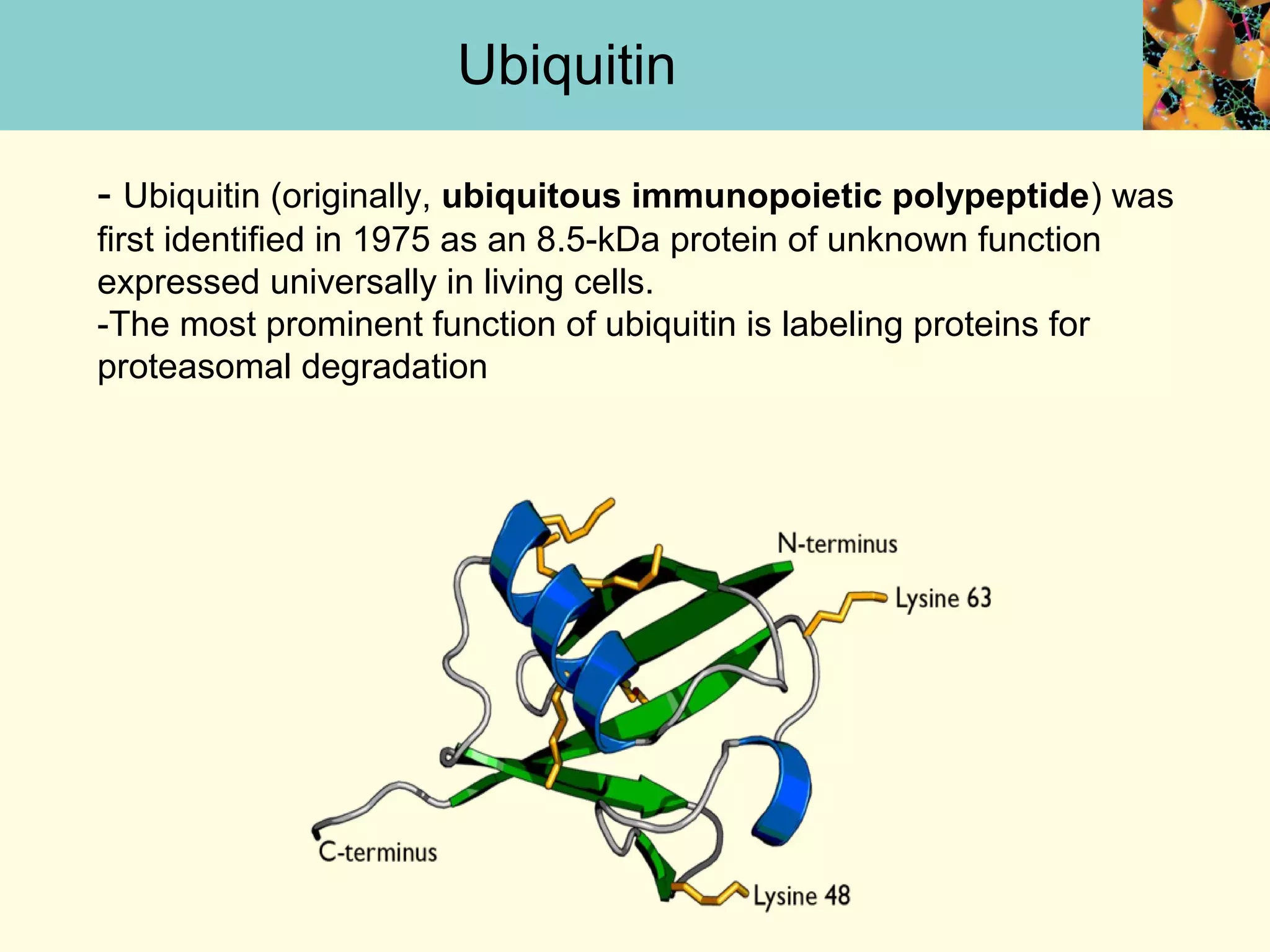
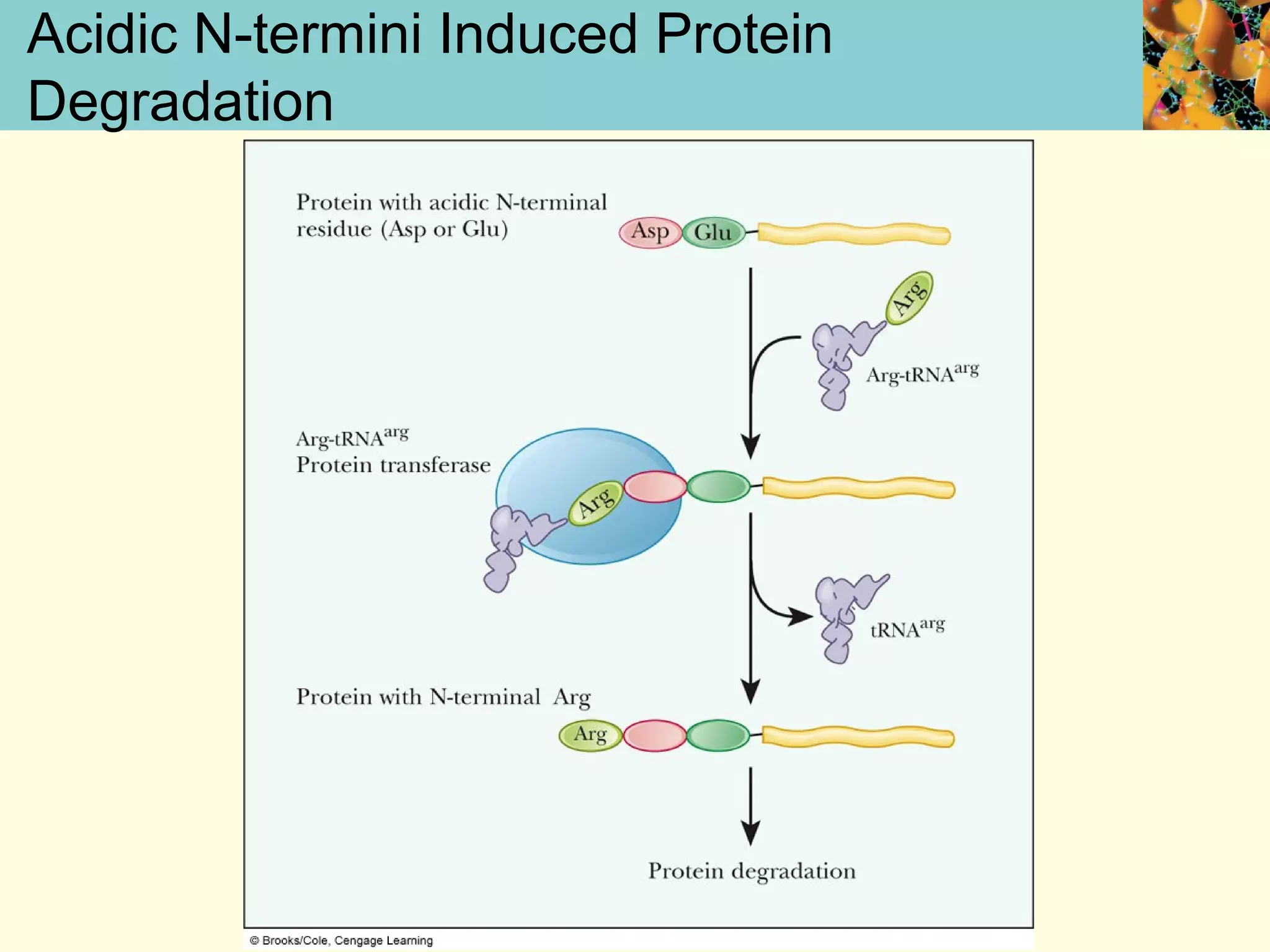
This document summarizes key aspects of protein synthesis, including translation of mRNA into a polypeptide chain. It discusses the genetic code and how triplet codons specify amino acids. The stages of translation - initiation, elongation, and termination - are described. Post-translational modifications and protein degradation are also covered. Protein synthesis requires various ribosomal and transfer RNA components to translate the genetic message into proteins.