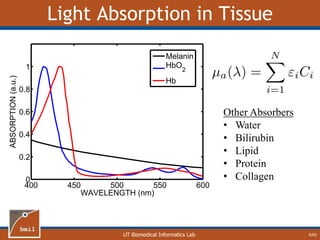
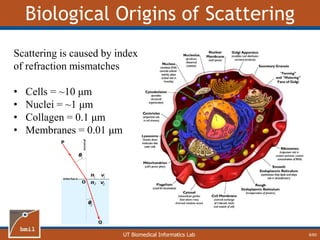
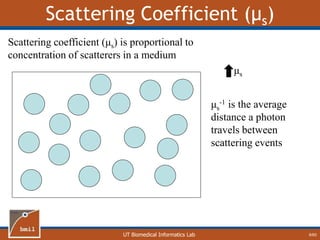
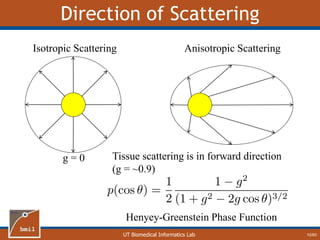
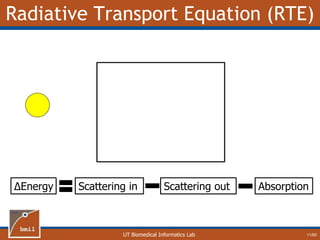
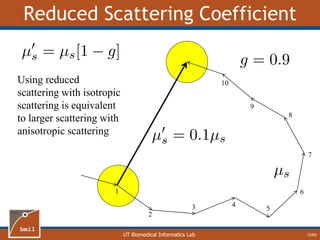
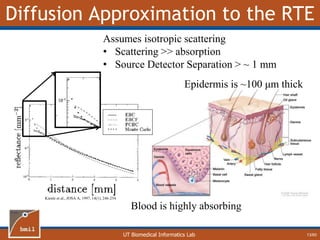
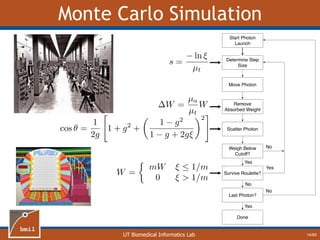
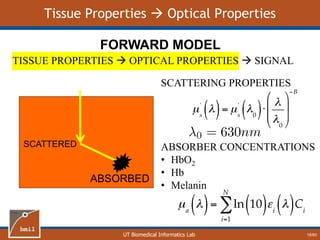
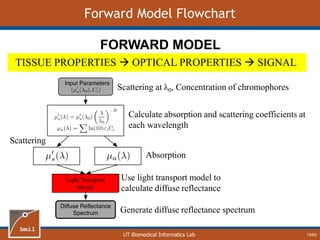
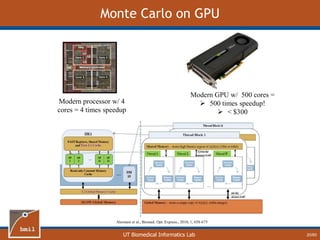
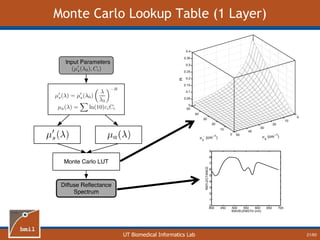
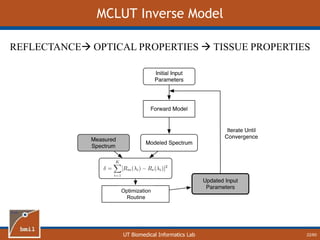
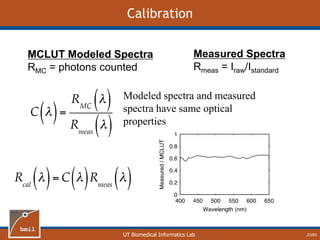
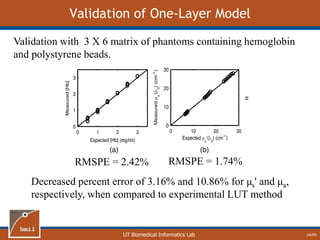
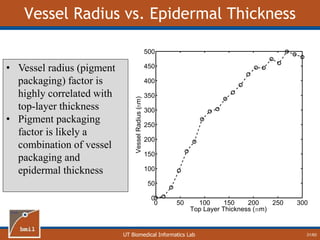
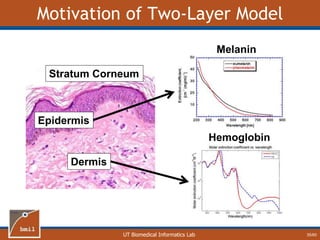
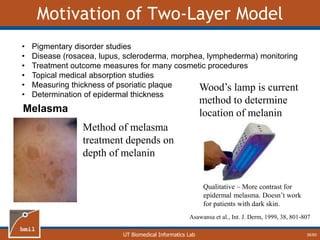
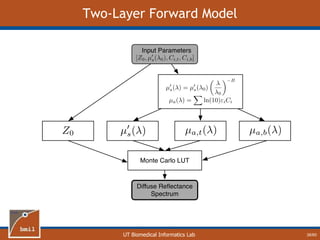
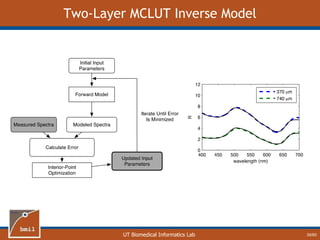
This document provides an overview of diffuse reflectance spectroscopy (DRS) and its applications in biomedical research. DRS uses non-invasive optical measurements to extract information about tissue optical properties and composition. It describes how light interacts with tissue through absorption and scattering. The document also discusses techniques for modeling light transport in tissue, such as Monte Carlo simulations and the development of lookup tables to relate tissue optical properties to DRS measurements. This allows DRS data to be analyzed to determine properties like hemoglobin concentration and oxygen saturation in tissue.







![UT Biomedical Informatics Lab
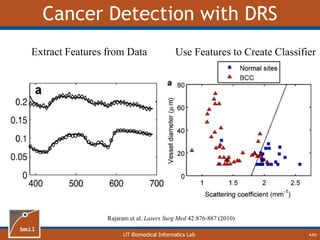
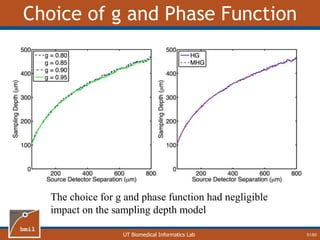
Fit Two-Layer Data with One Layer Model
26/60
Hb + HbO2
melanin
melanin +
Hb + HbO2
1. Create two-layer
spectra
2. Fit with one-
layer model
• [mel]
• [Hb]
• SO2
• Scattering
• Epidermal thickness
• [mel]
• [Hb]
• SO2
• Scattering
• Vessel radius
Notice that the fit
is very good](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-27-320.jpg)
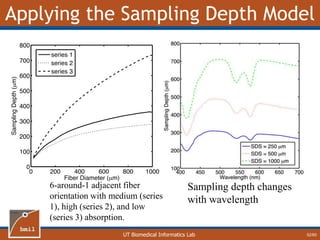
![UT Biomedical Informatics Lab
Melanin
27/60
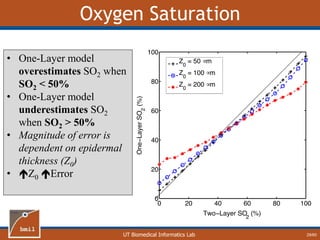
• One-Layer model
underestimates [mel]
• Magnitude of error is
dependent on epidermal
thickness (Z0)
• Z0 Error](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-28-320.jpg)
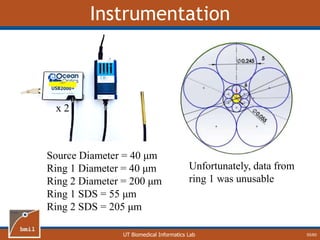
![UT Biomedical Informatics Lab
Hemoglobin
28/60
• One-Layer model
underestimates [Hb]
• Magnitude of error is
dependent on epidermal
thickness (Z0)
• Z0 Error](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-29-320.jpg)

![UT Biomedical Informatics Lab
Correlation Between [Hb] and [mel]
32/60
R = 0.04 R = 0.80
One-layer assumption causes artificial correlation
between [Hb] and [mel]](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-33-320.jpg)
![UT Biomedical Informatics Lab
Conclusions about One-Layer Errors
Causes underestimation of [Hb] and [mel]
– Magnitude of error is function of epidermal
thickness
Causes error in SO2 that is a function of
epidermal thickness as well as SO2
Vessel Packaging factor and epidermal
thickness are highly correlated
Causes an artificial correlation in [Hb] and
[mel]
33/60](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-34-320.jpg)


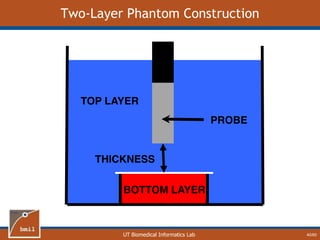
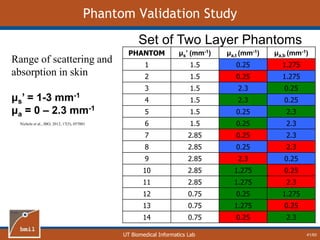
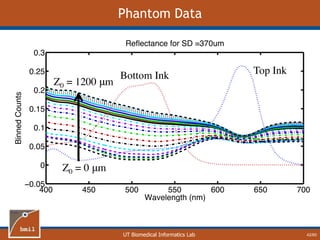
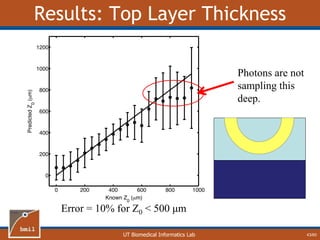
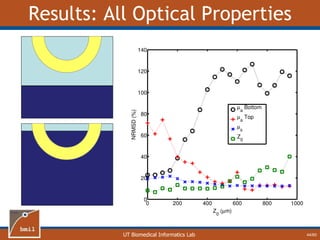
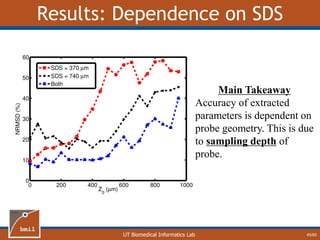

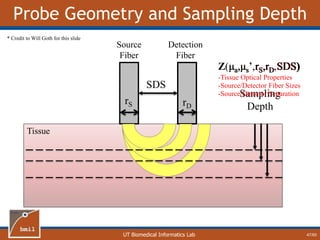
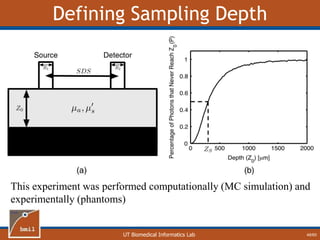
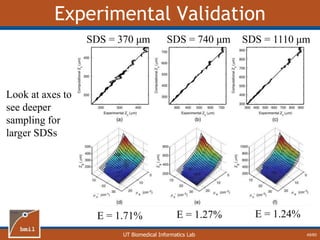
![UT Biomedical Informatics Lab
Analytical Model of Sampling Depth
50/60
This expression can be
used to aid in the design
of application specific
DRS probesE = 2.89%
Expression was found using
TableCurve 3D. Free parameters
[a1, a2, a3, a4] were selected using a
least-squares fitting algorithm.](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-51-320.jpg)


![UT Biomedical Informatics Lab
Melanin
56/60
Back Calf Cheek Forearm Palm
0
0.5
1
1.5
2
2.5
3
3.5
4
MelaninConcentration(mg/ml)
Palm has less melanin,
which agrees with the
expected result.
Average of 1.83 mg/ml
is within range of
published values for
melanin concentration
[0-5 mg/ml]](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-57-320.jpg)
![UT Biomedical Informatics Lab
Hemoglobin
57/60
Back Calf Cheek Forearm Palm
0
0.5
1
1.5
2
2.5
3
3.5
HemoglobinConcentration(mg/ml)
Higher levels of
hemoglobin in the face
and forearm agrees
with the expected
results.
Average of 1.37 mg/ml
is within range of
published values for
[Hb]
[0.5-10 mg/ml]](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-58-320.jpg)
![UT Biomedical Informatics Lab
Scattering
58/60
No significant
difference between
anatomical locations.
Average of 22.75 cm-1
is within range of
published values for
scattering at 630 nm
[15 – 25 cm-1]](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-59-320.jpg)
![UT Biomedical Informatics Lab
Epidermal Thickness
59/60
Back Calf Cheek Forearm Palm
0
20
40
60
80
100
120
140
160
EpidermalThickness(mm)
No significant
difference between
anatomical locations.
Average of 90 μm is
within range of
published values for
epidermal thickness.
[40 – 200 μm]
We expected to see a
difference between
anatomical locations.](https://image.slidesharecdn.com/defense-160707152605/85/Bio-optical-Sensing-Dissertation-60-320.jpg)