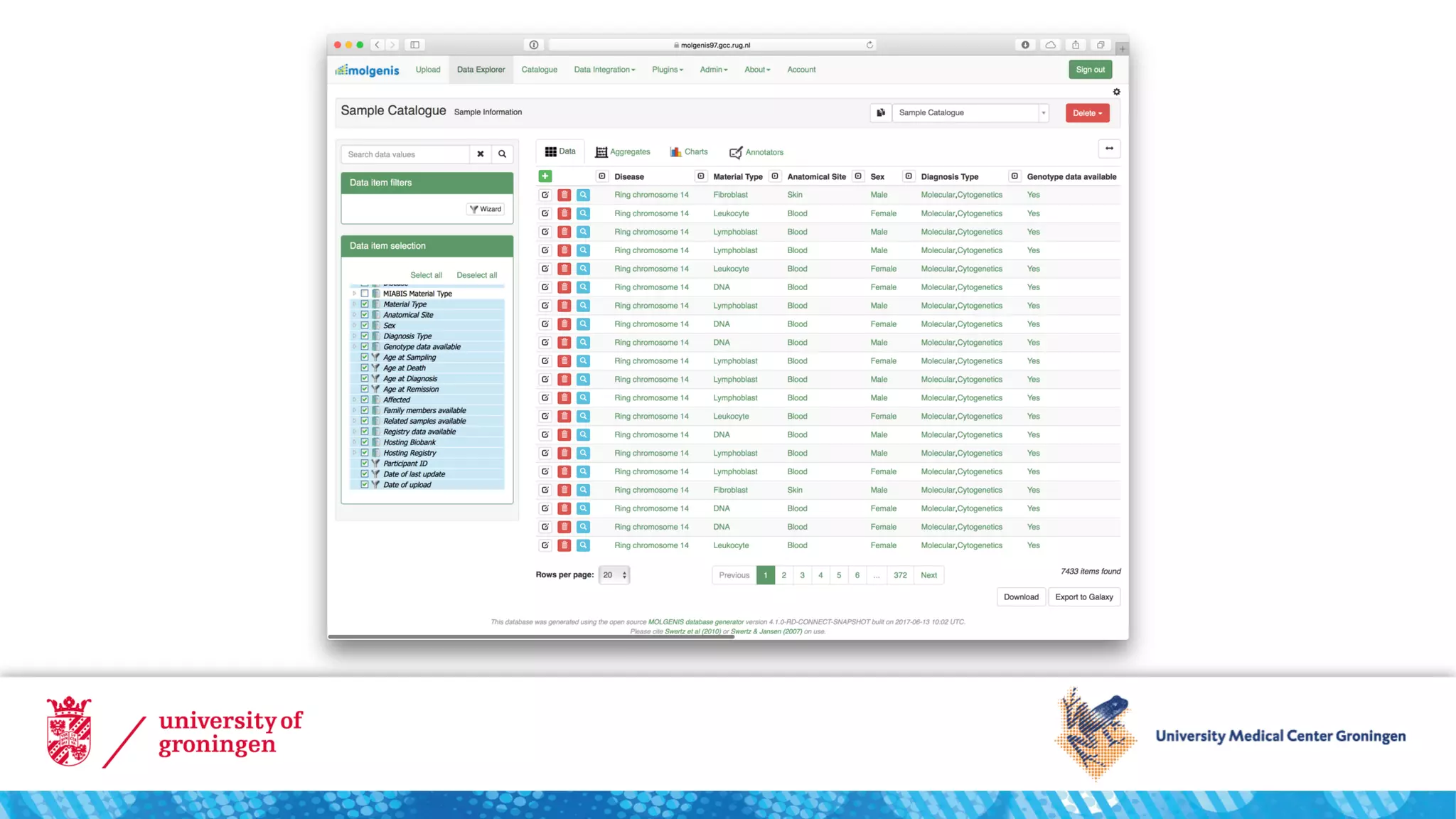
This document discusses making biobank data and samples FAIR (Findable, Accessible, Interoperable, and Reusable).
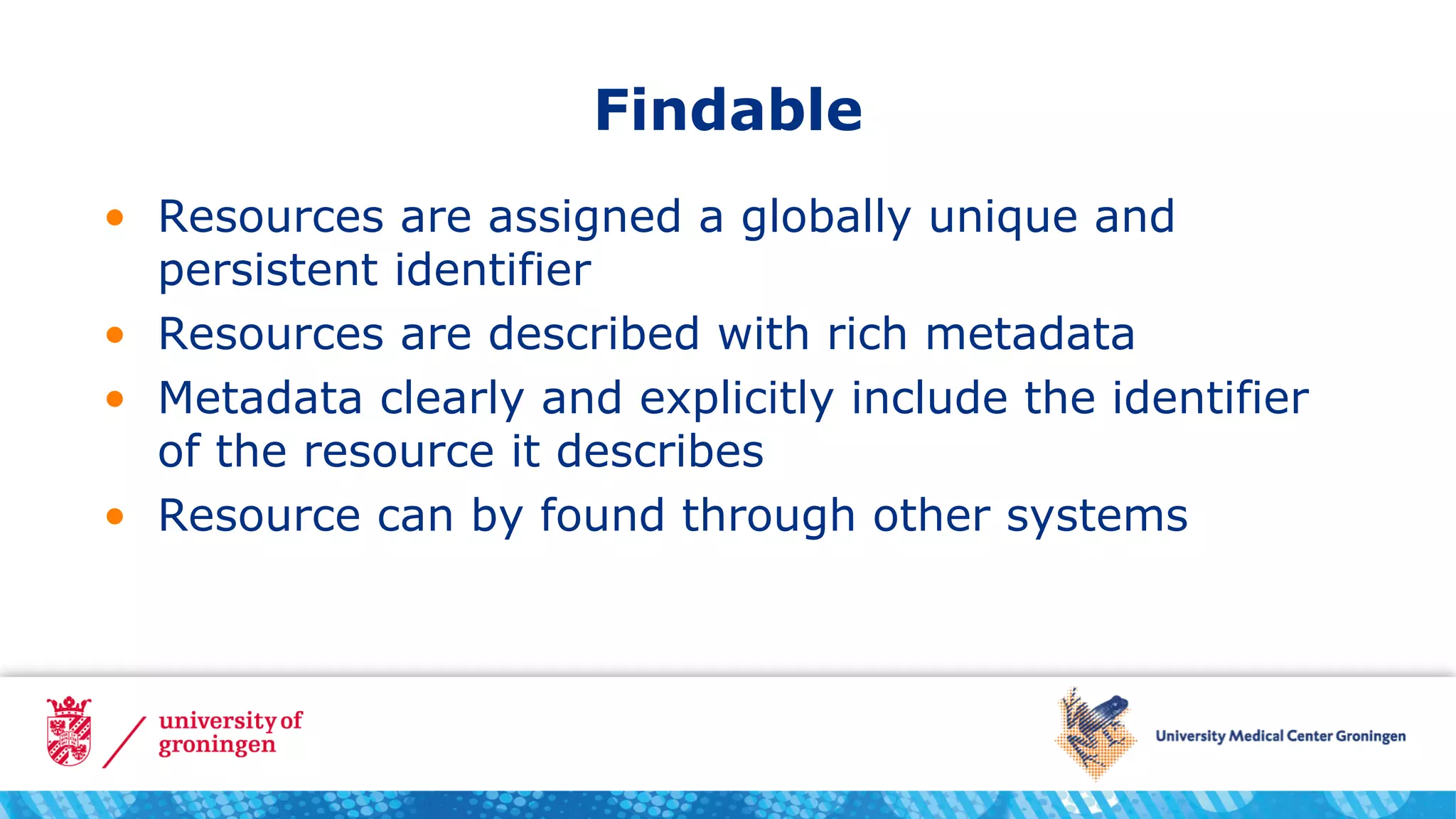
It explains the four FAIR principles and provides examples of how to apply each one. To make resources findable, they need unique and persistent identifiers, rich metadata, and to be discoverable through other systems. To make them accessible, they need to be retrievable using open standards. To make them interoperable, standards for knowledge representation like ontologies should be used. And to make them reusable, they need to be richly described and released with clear usage terms and provenance.
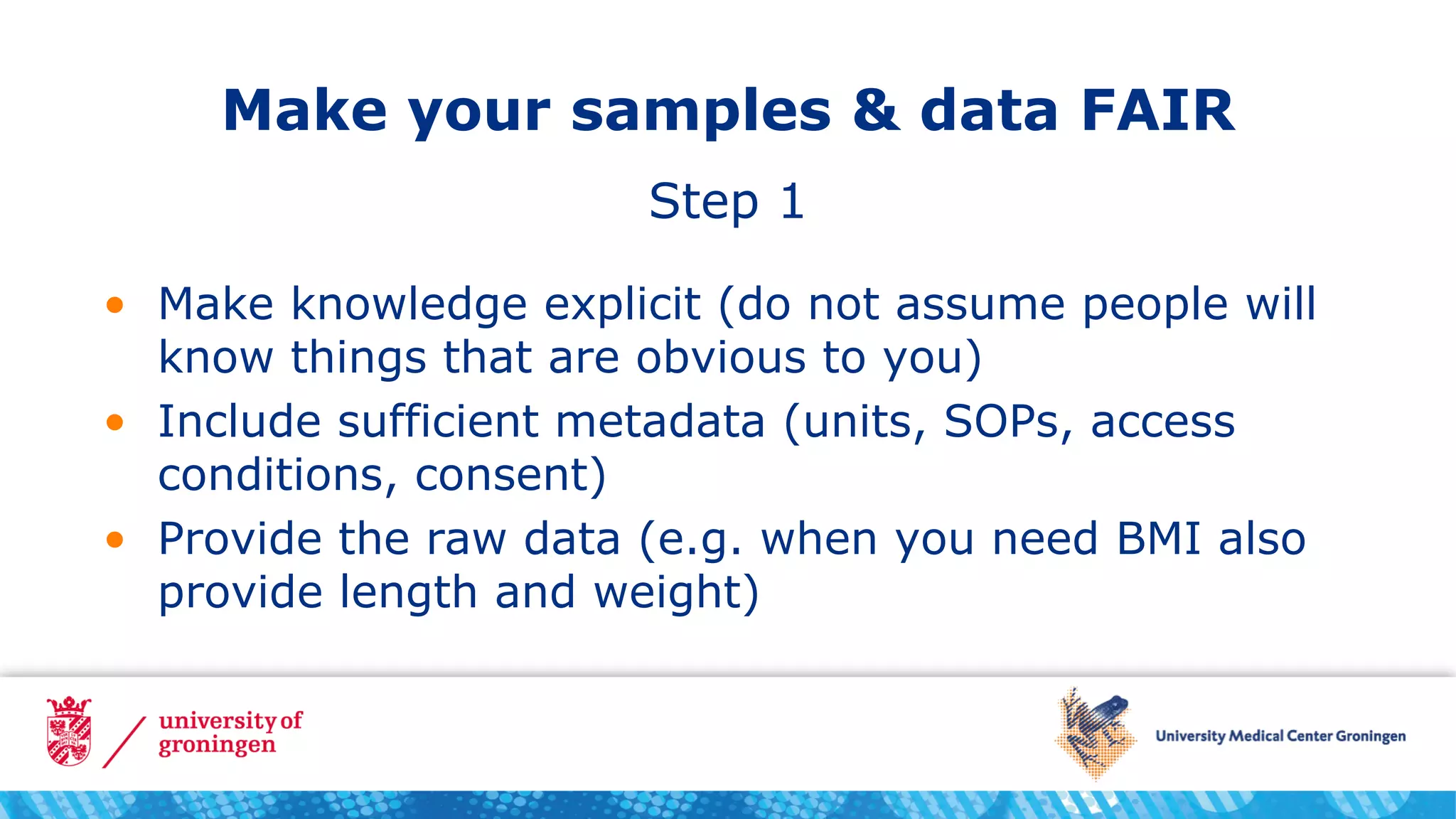
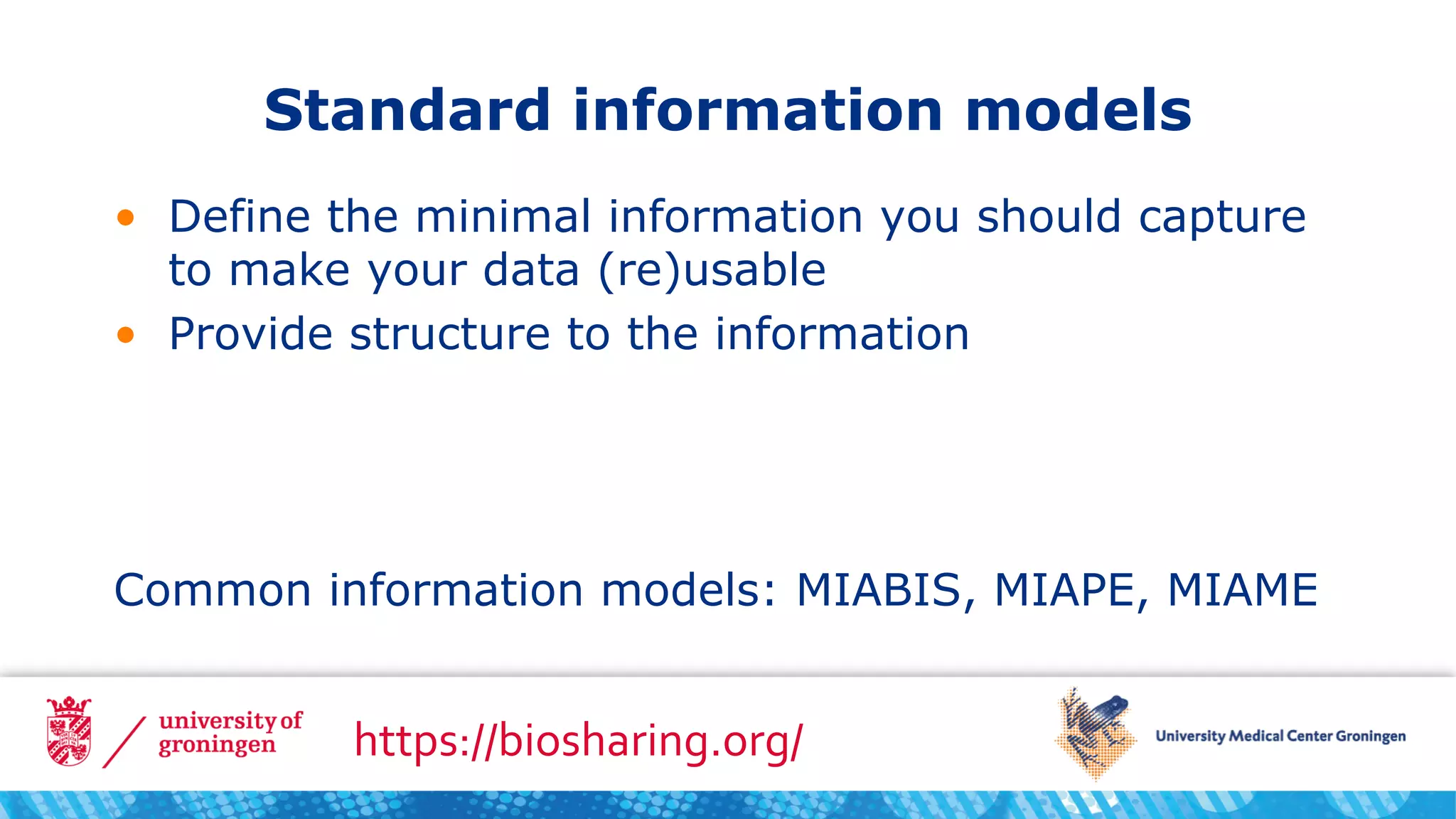
The document recommends three steps to make samples and data FAIR: include sufficient metadata using