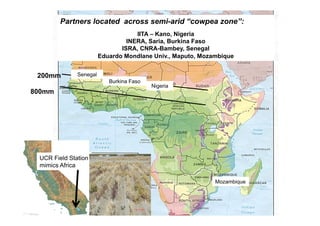
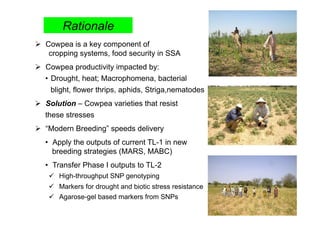
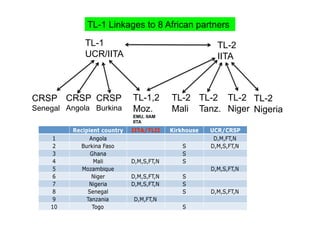
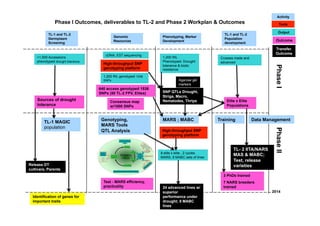
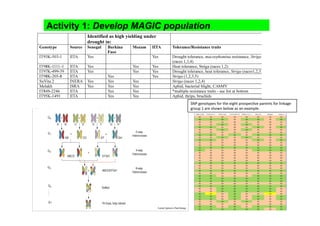
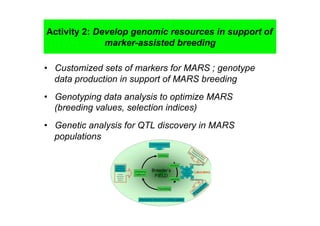
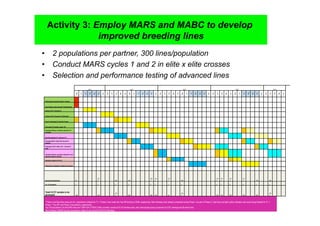
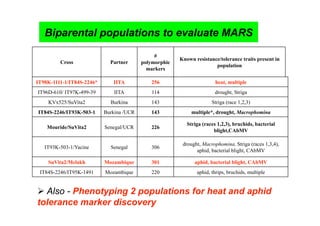
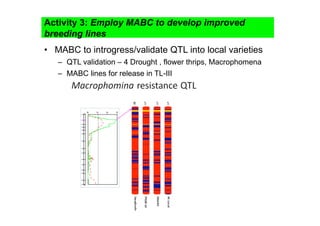
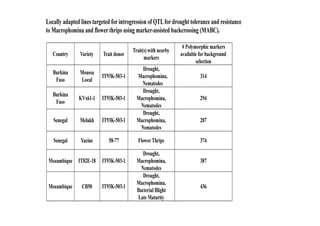
This document outlines a draft workplan for improving cowpea productivity in marginal environments in Sub-Saharan Africa from November 2009. It involves partners across the cowpea growing region using modern breeding strategies like MARS and MABC. The workplan builds on outputs from Phase 1 including drought tolerant germplasm, markers for stress resistance, and a high density SNP map. Key activities include developing a MAGIC population, genomic resources to support marker-assisted breeding, and employing MARS and MABC over two cycles to develop improved cowpea varieties for release. The goal is to speed delivery of cowpea varieties resistant to drought, diseases and pests to improve food security.