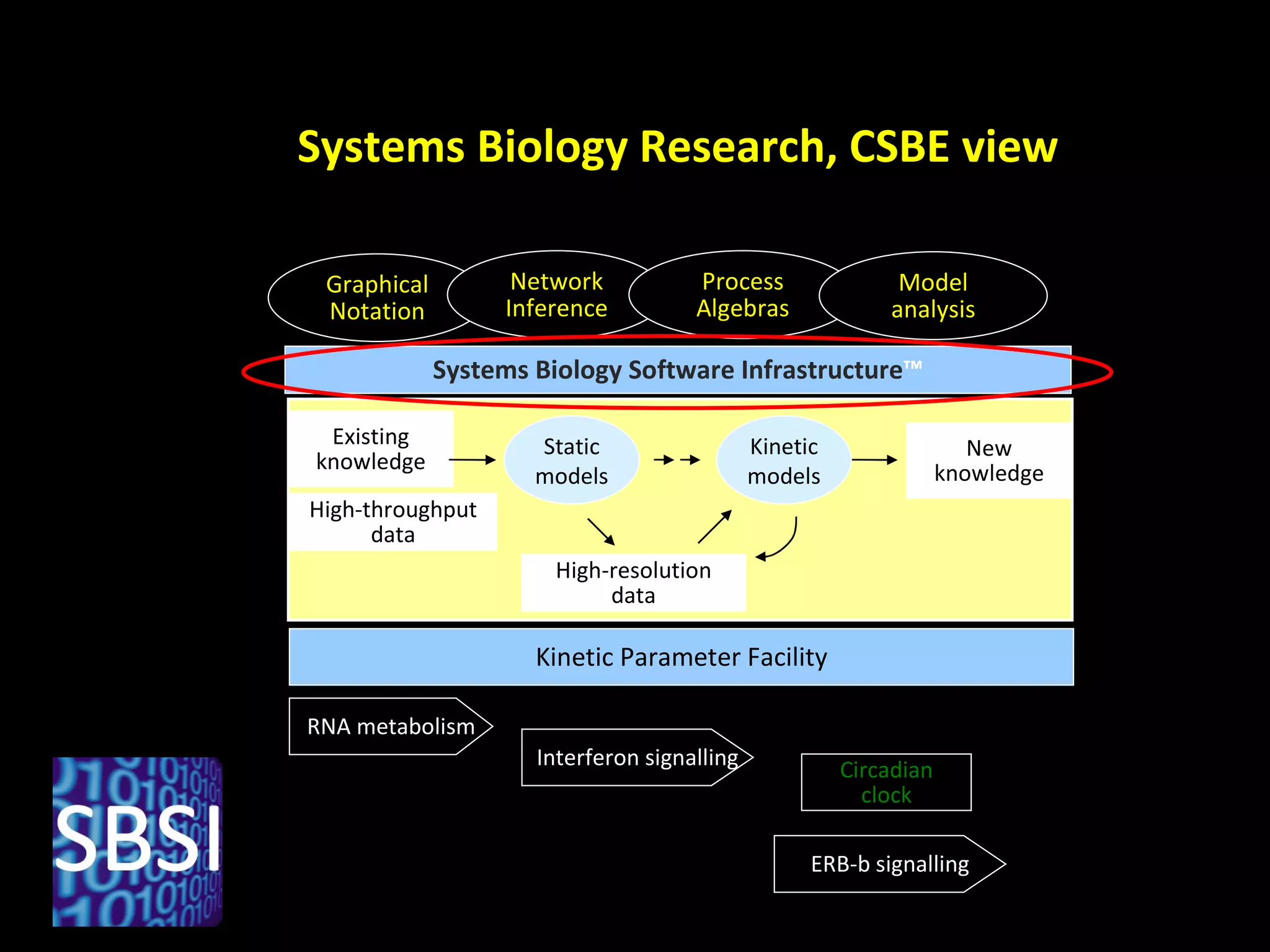
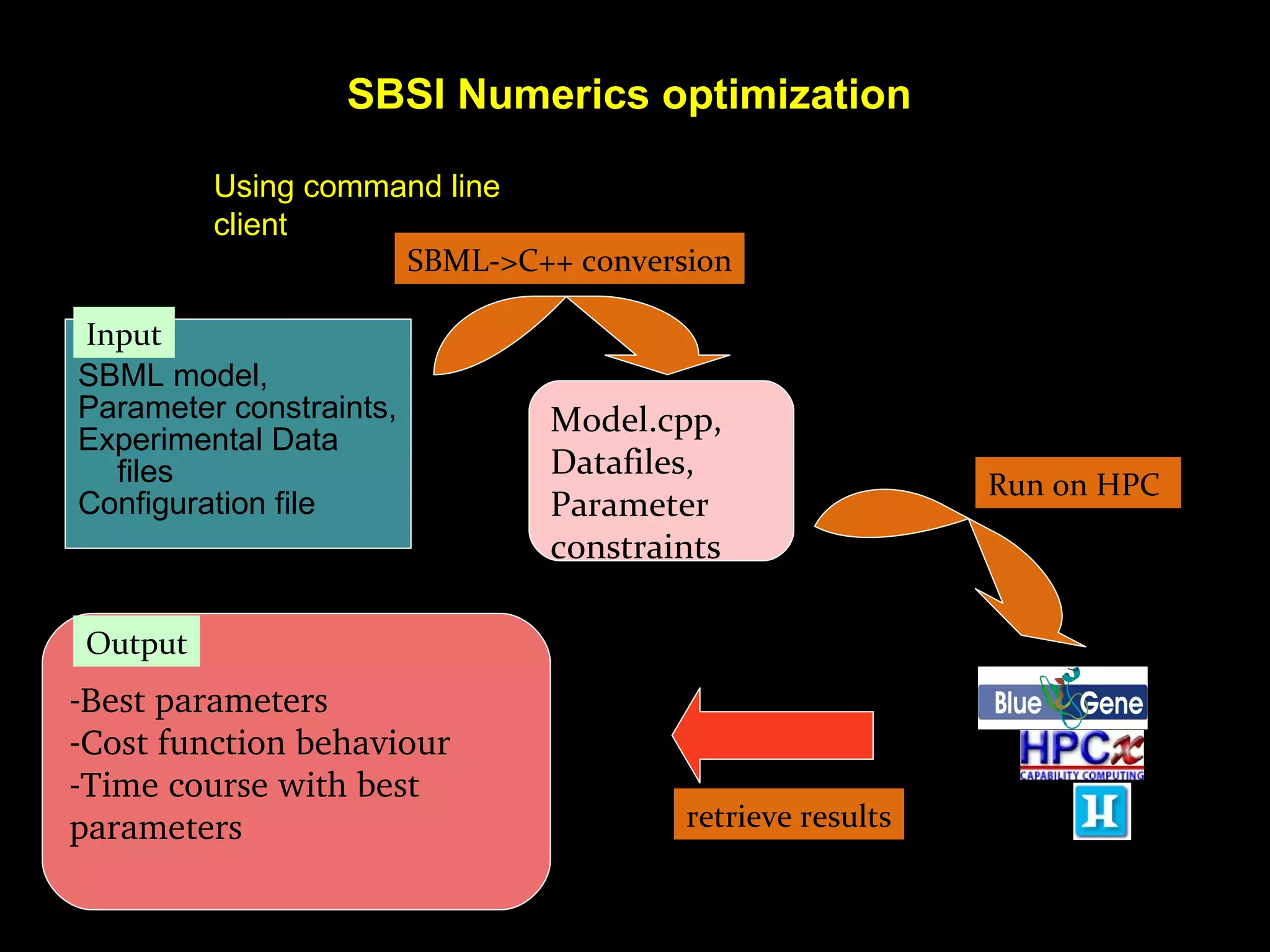
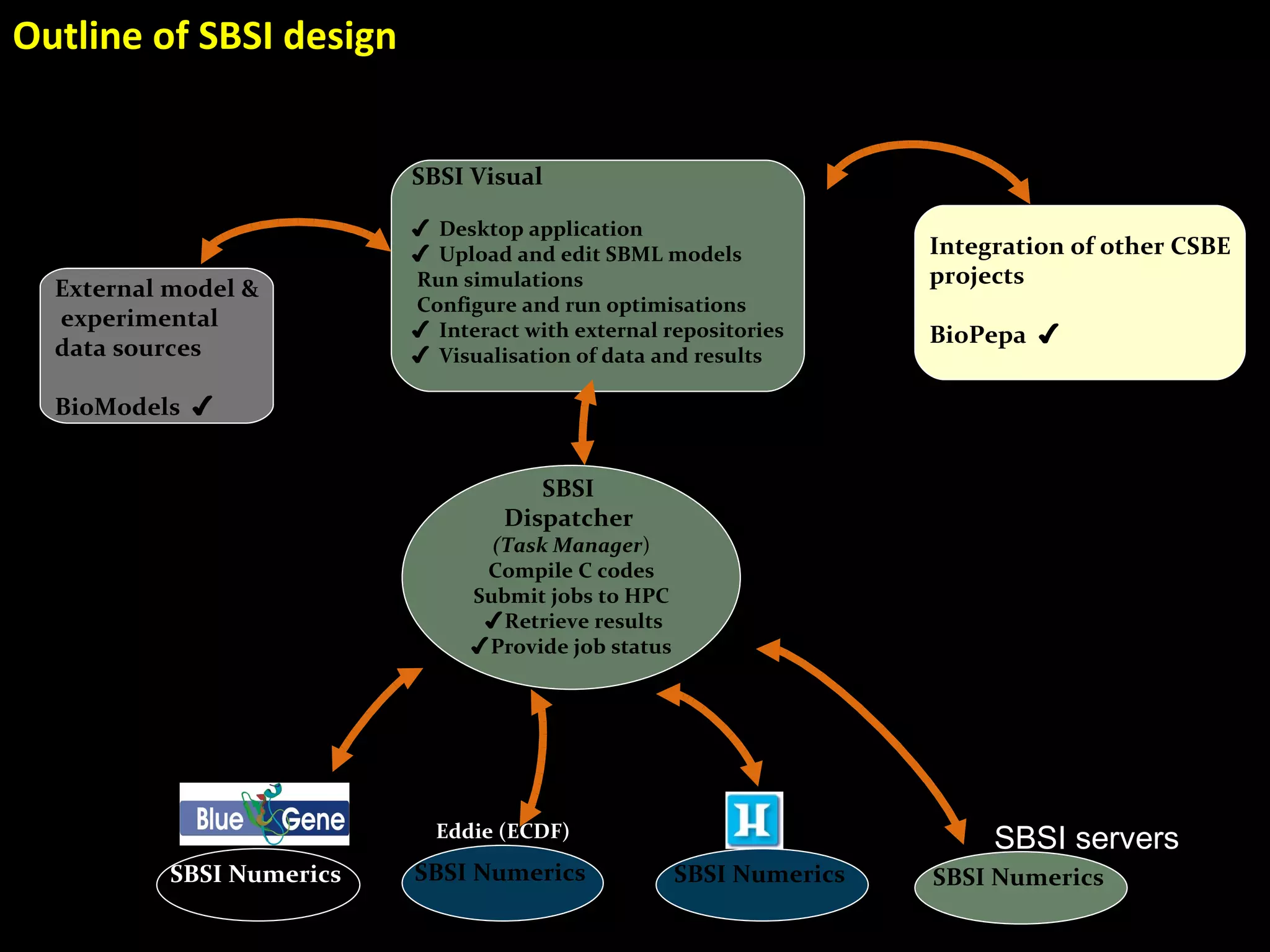
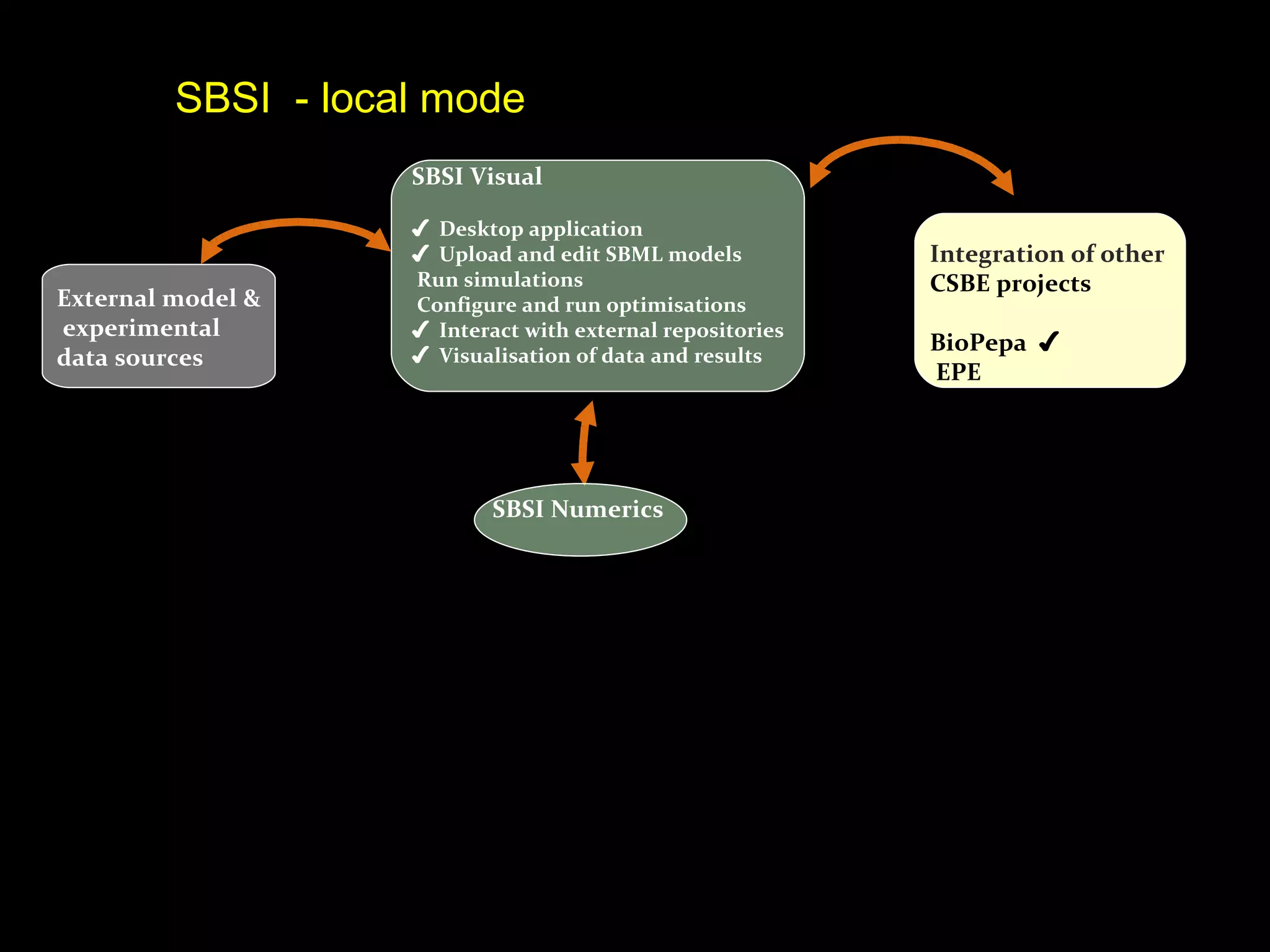
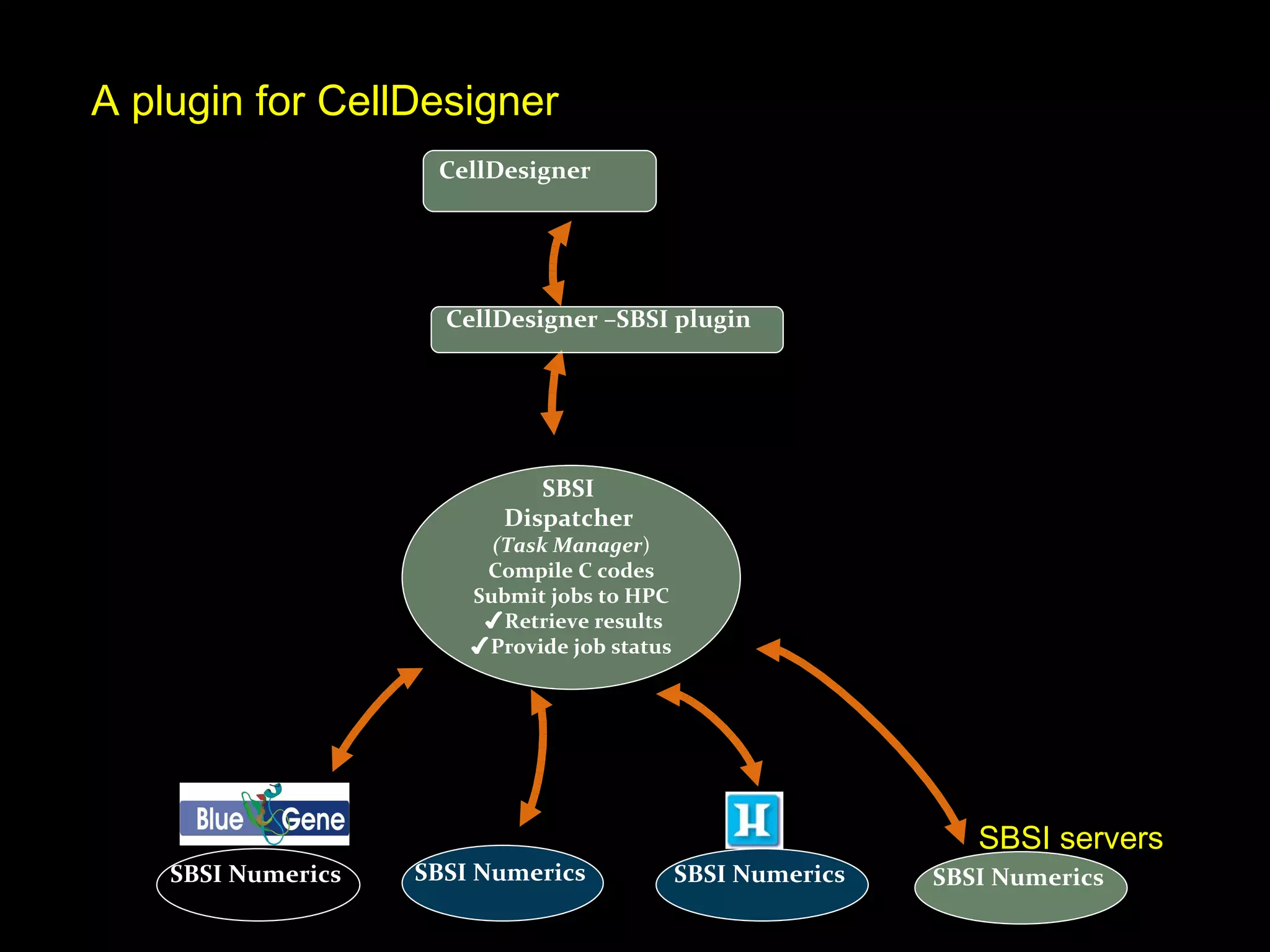
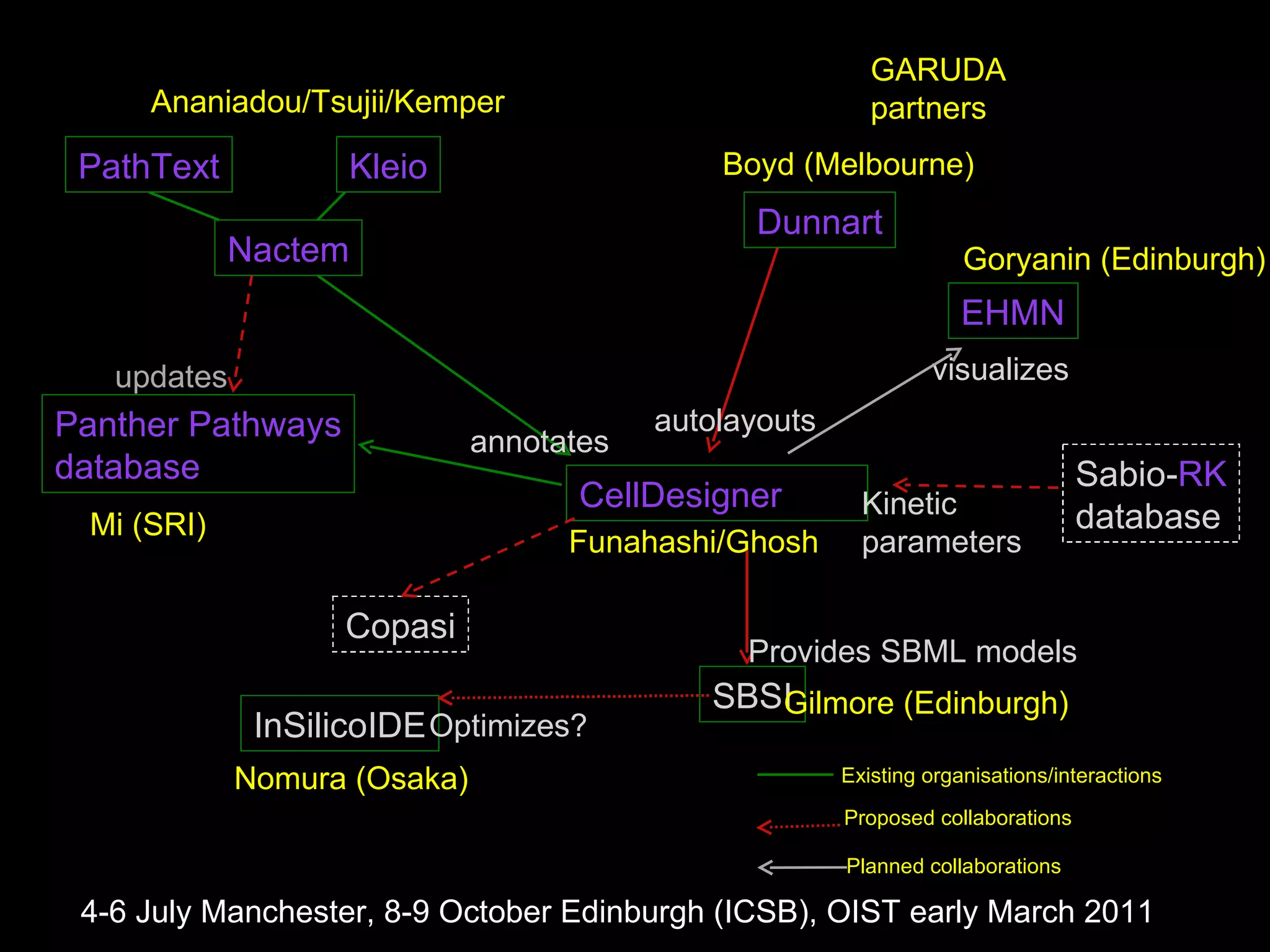
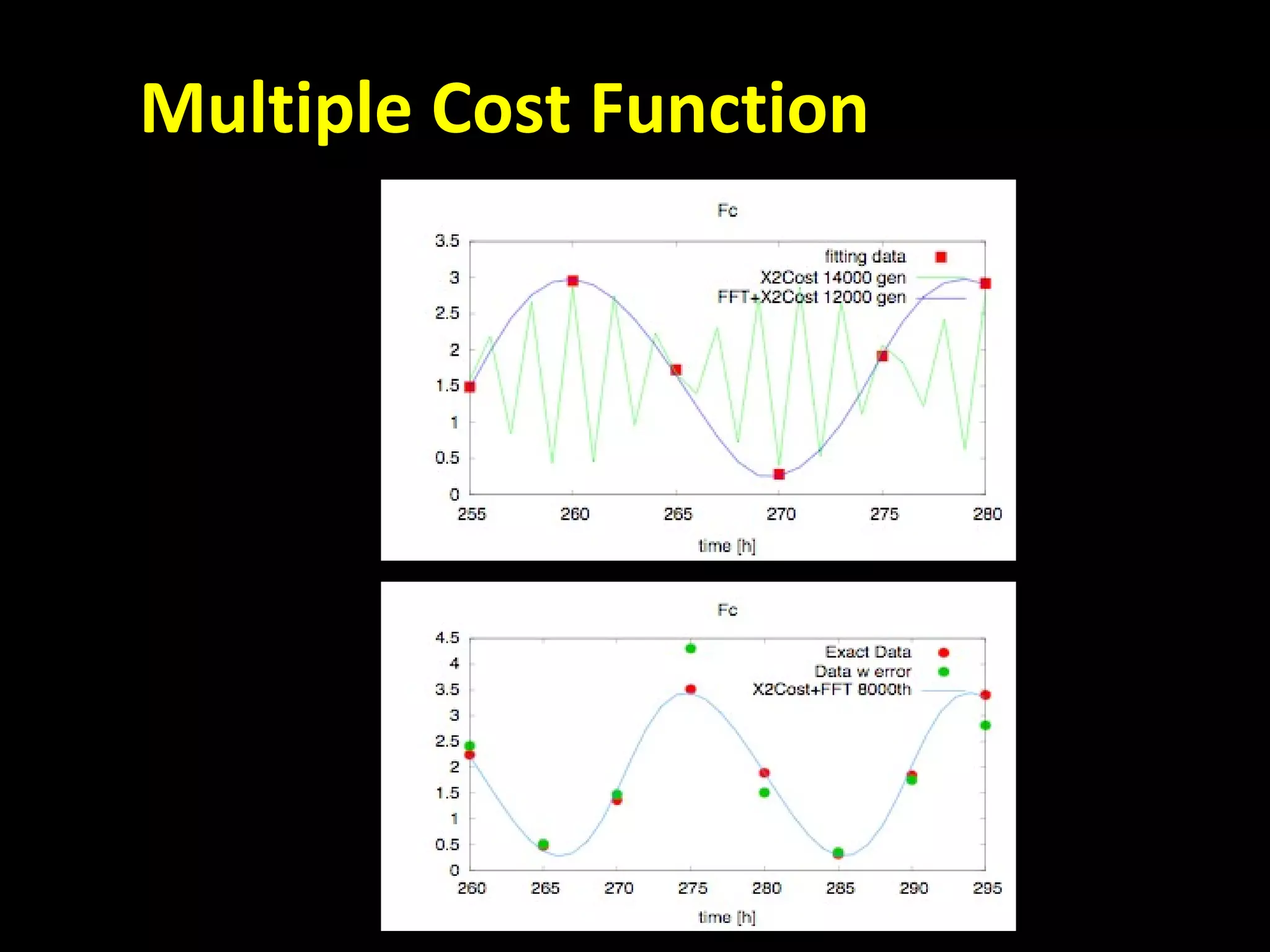
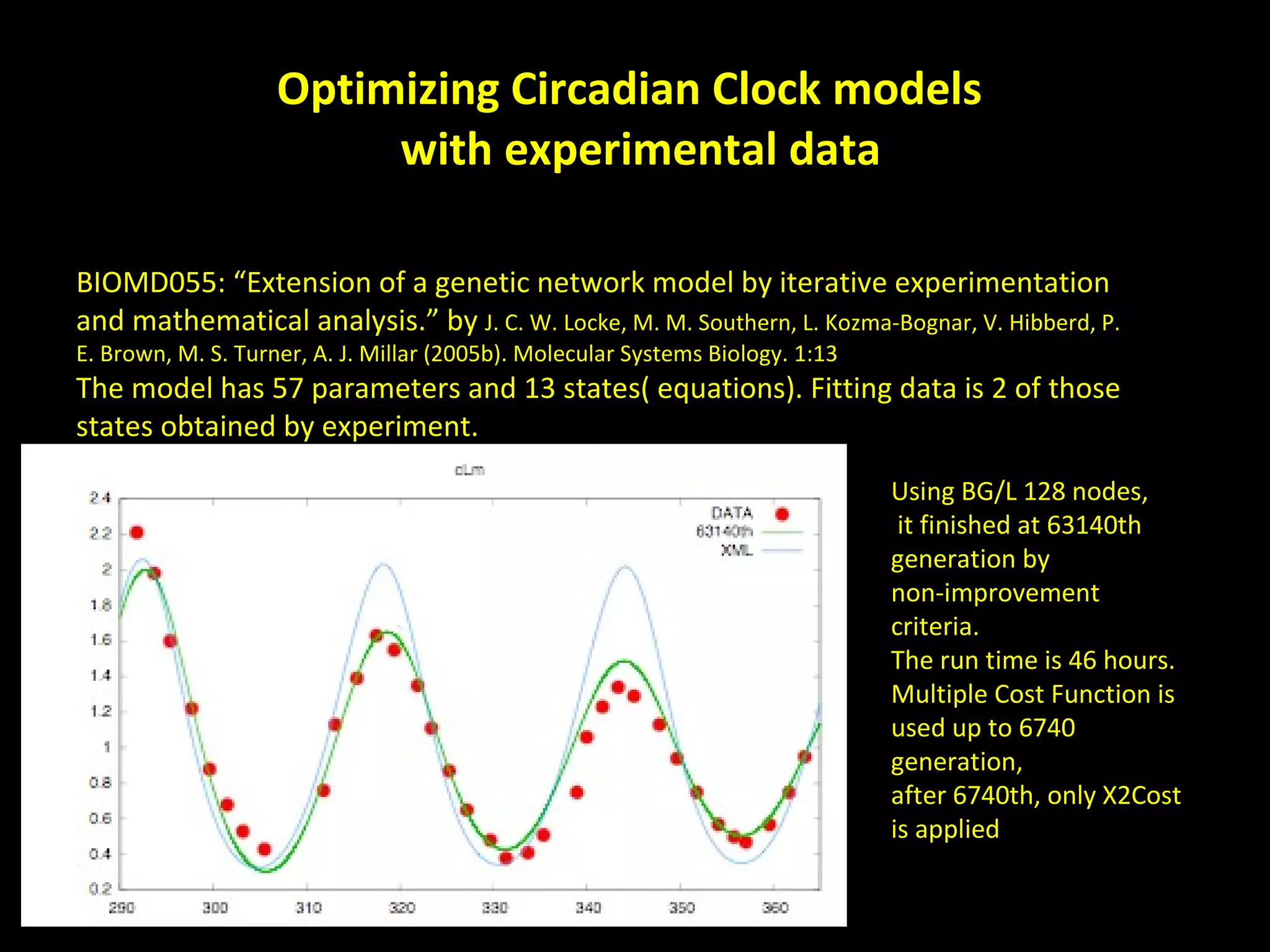
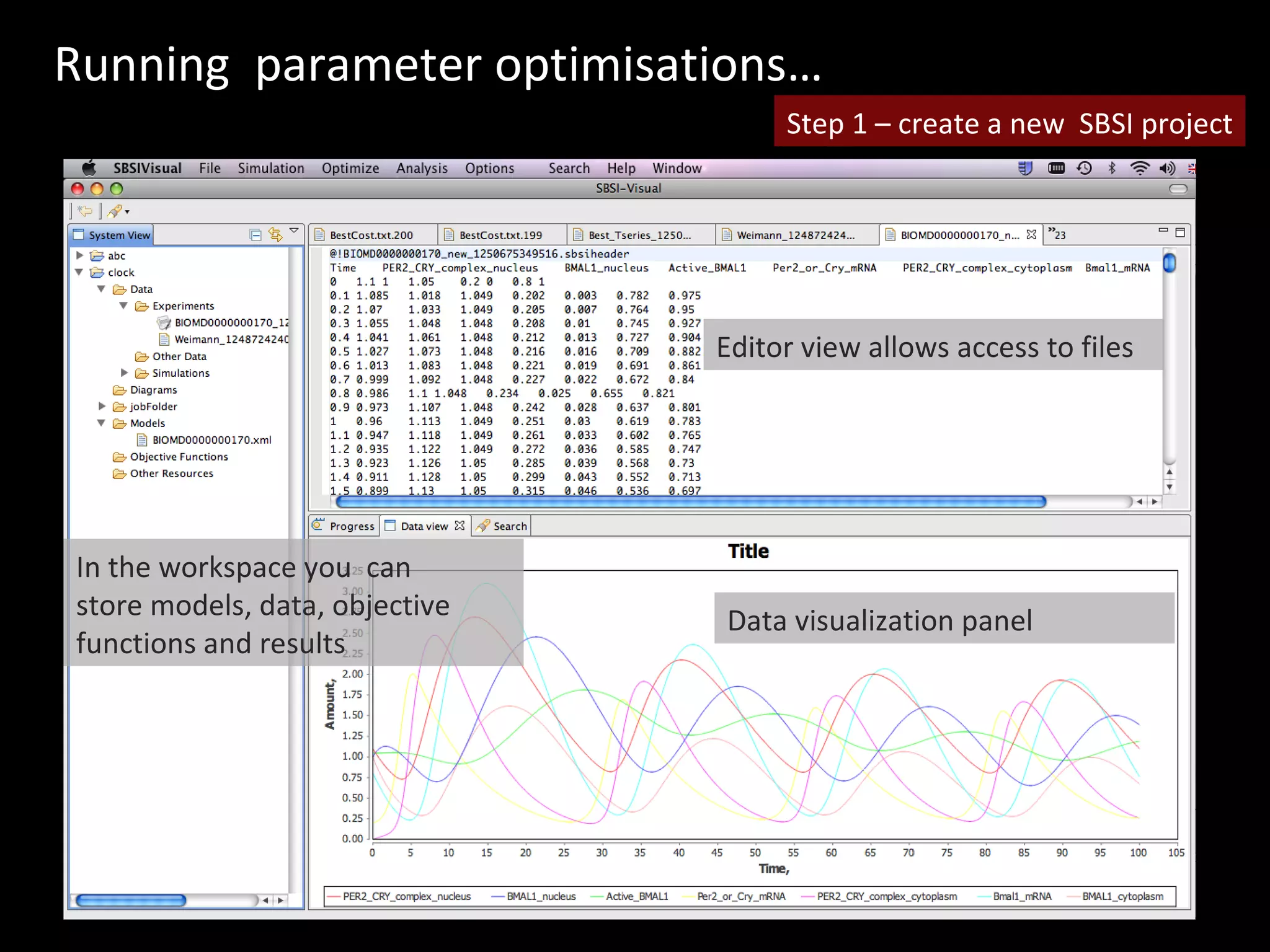
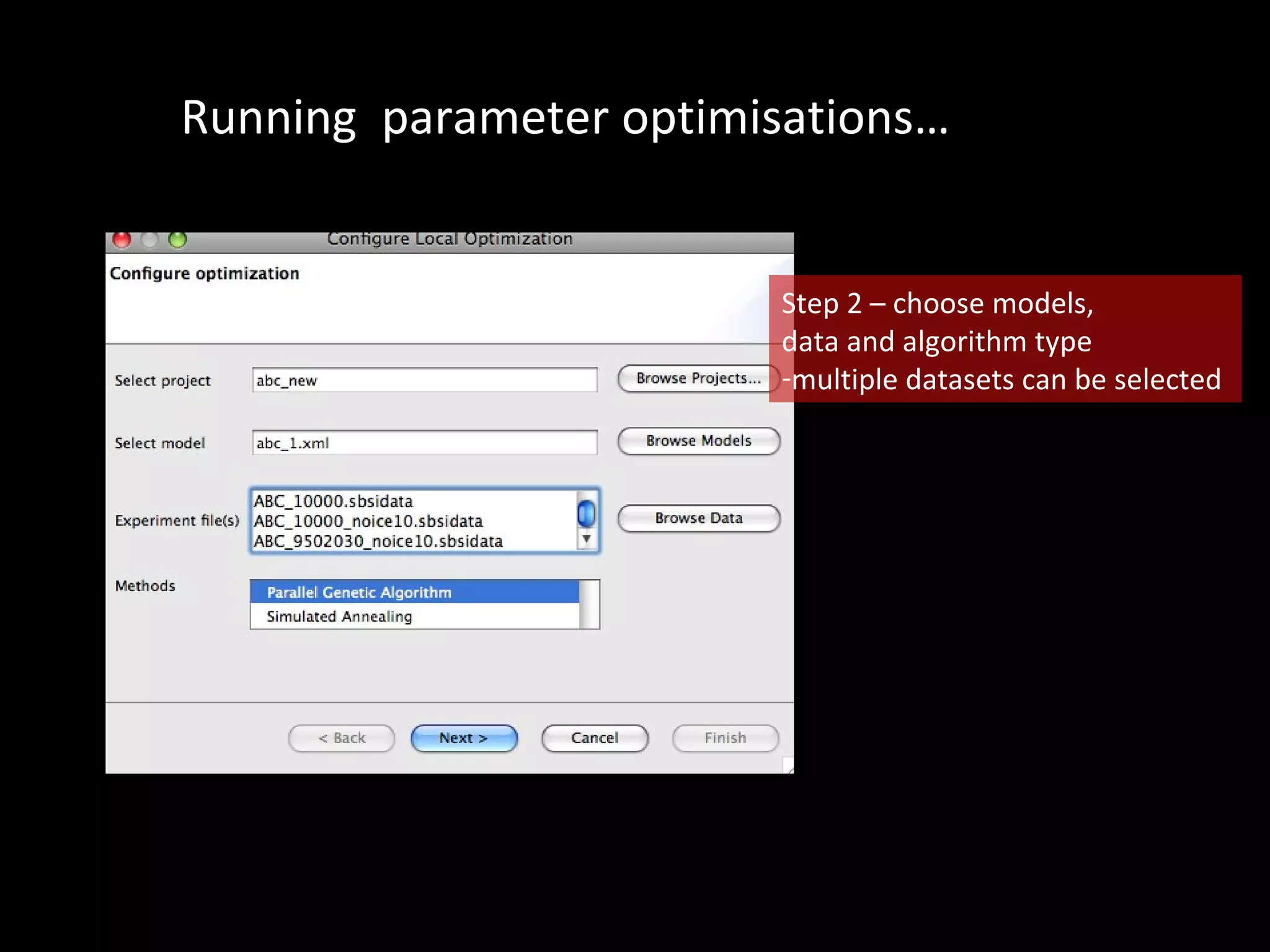
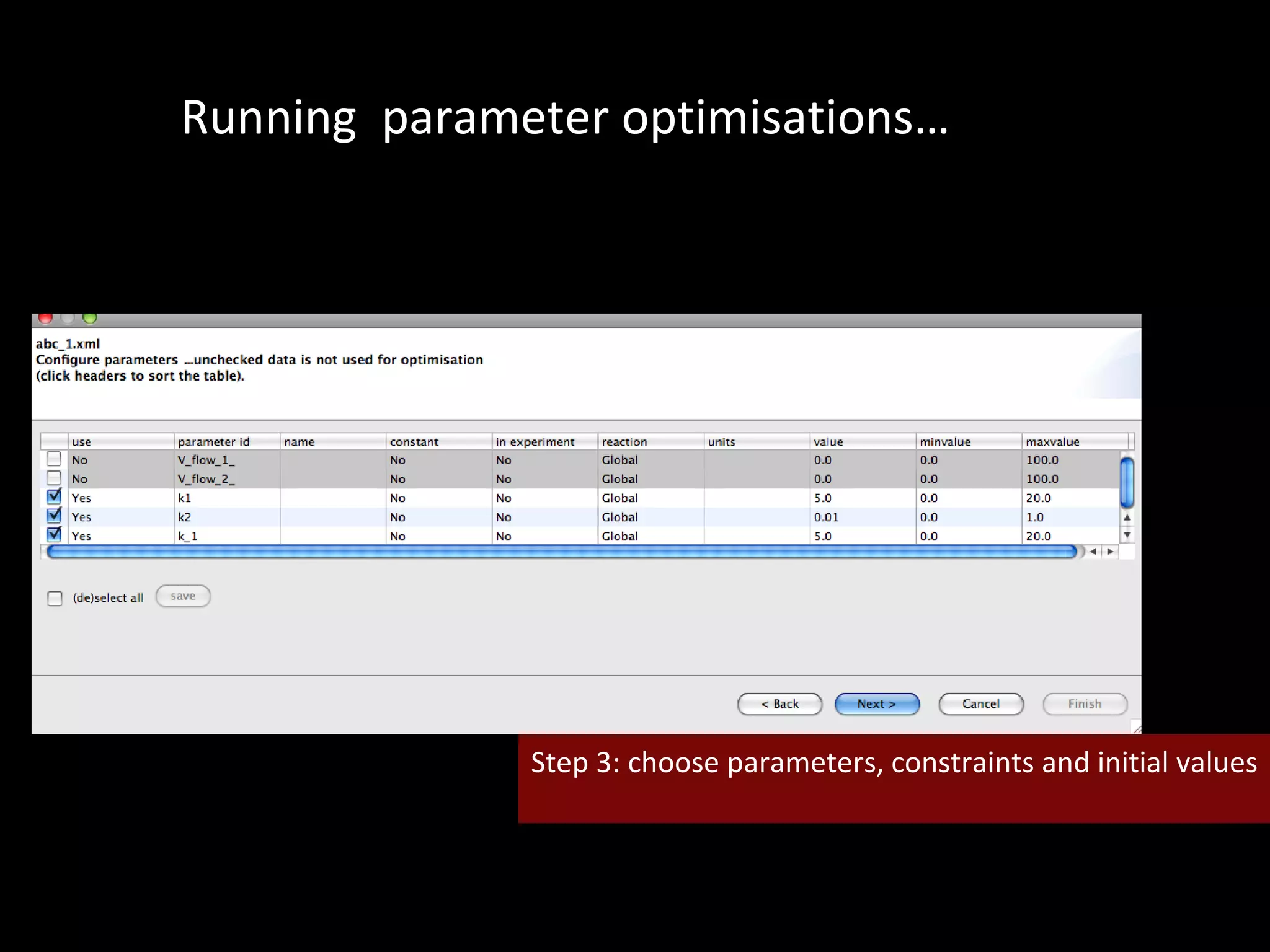
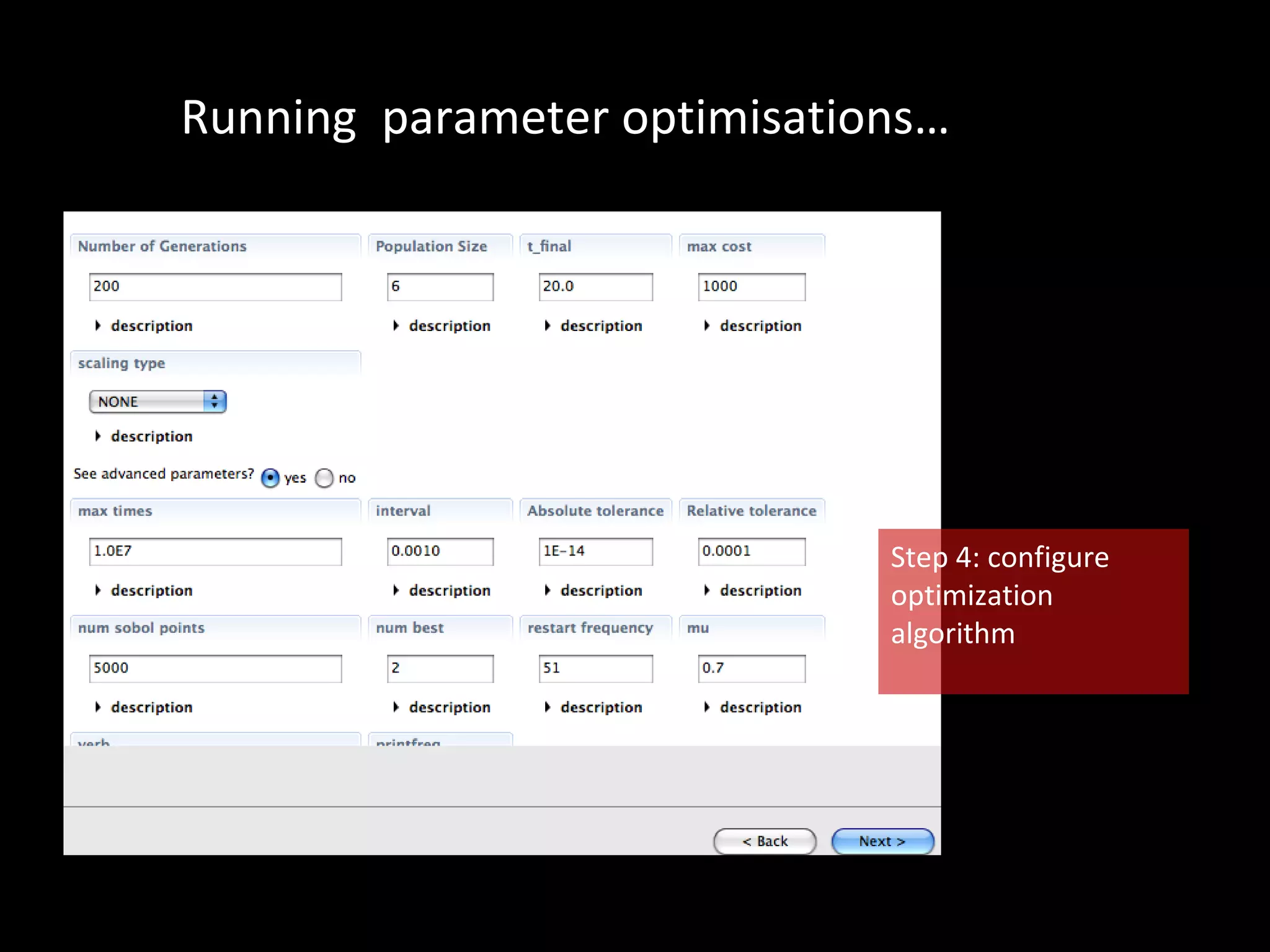
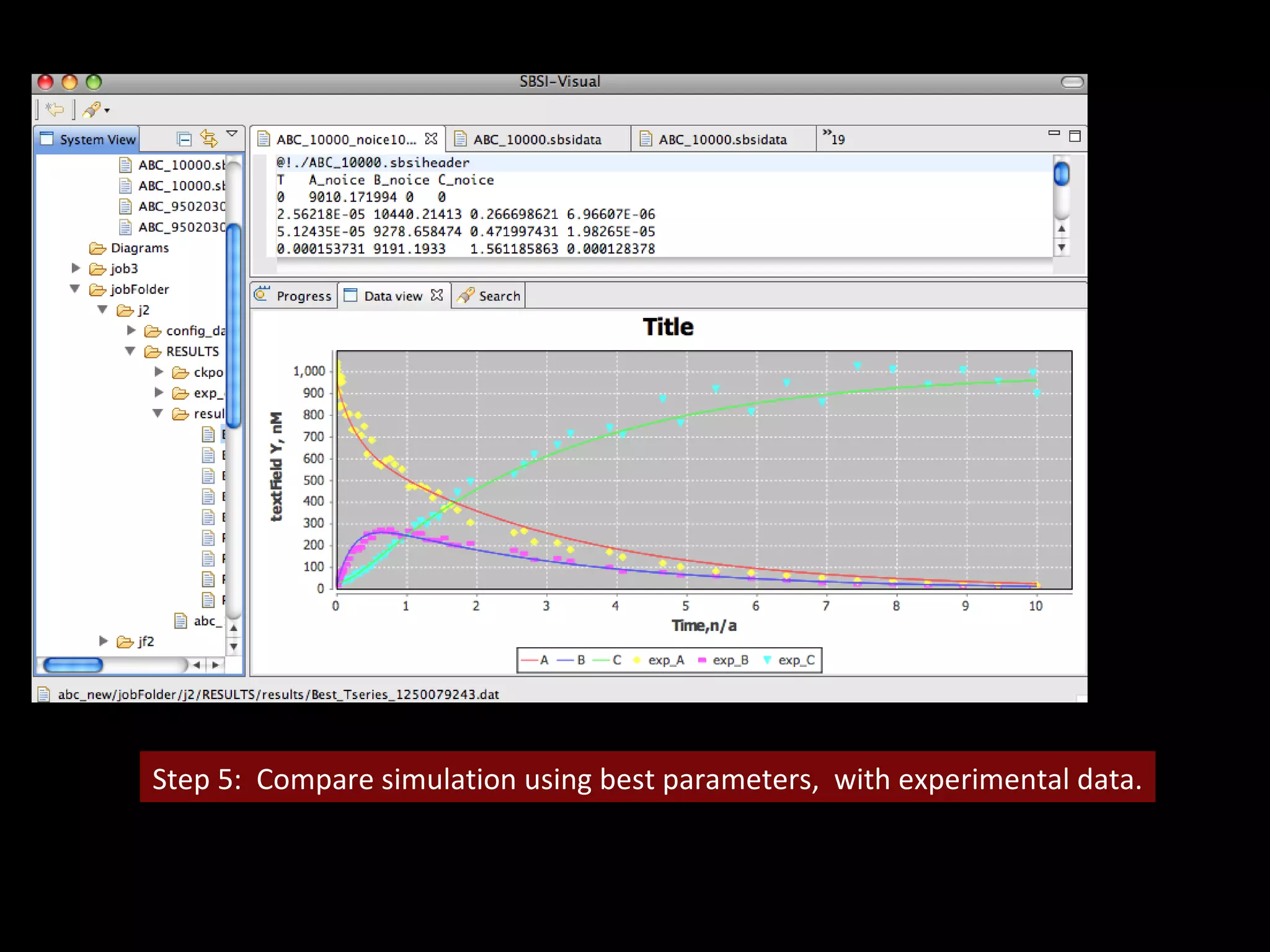
The document discusses the Systems Biology Software Infrastructure (SBSI) project. SBSI aims to streamline the connection between biological data, models, and analysis. It allows for updating large-scale models and data with reduced overhead. The initial use case is parameter estimation for circadian clock models. SBSI includes tools for editing SBML models, running simulations, performing parameter optimizations, and visualizing results. It will integrate with other systems biology software and be accessible through the modeling tool CellDesigner. The goals for 2010 include releasing the SBSI code, providing access via Hector supercomputing, and growing the user base.