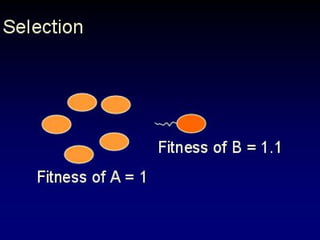
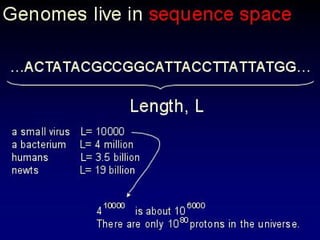
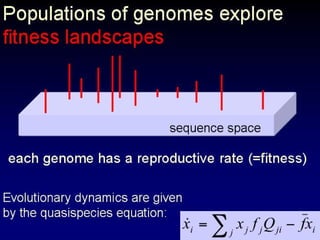
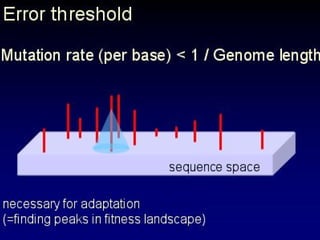
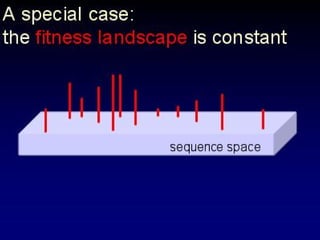
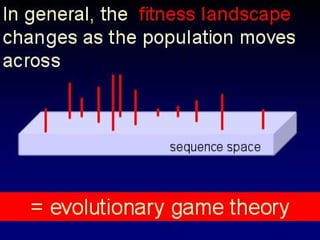
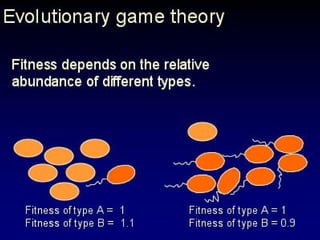
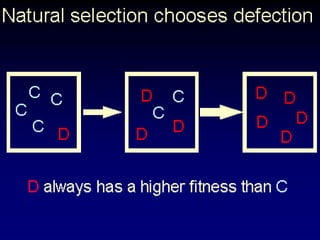
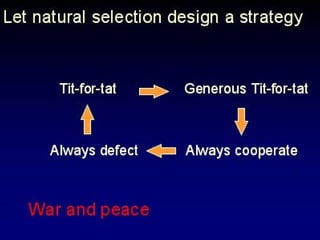
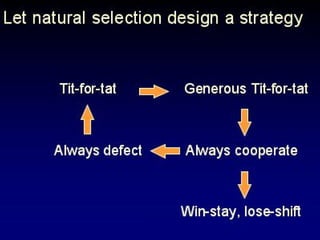
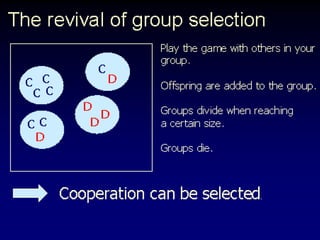
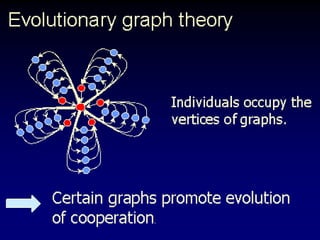
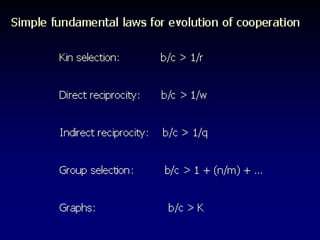
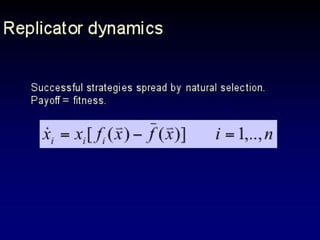
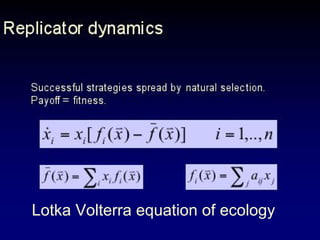
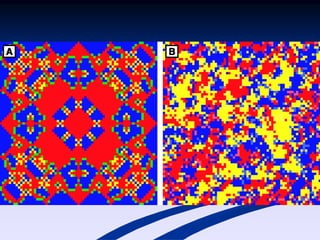
The document discusses the importance of understanding the design principles of metabolic networks and the evolutionary origins of organelles like mitochondria through the endosymbiont theory. It highlights the role of modularity in cellular functionality and the integration of complex systems, while emphasizing current research methods and ongoing investigations in the field. Additionally, it mentions a presentation on evolutionary dynamics, reflecting on concepts in evolutionary game theory.