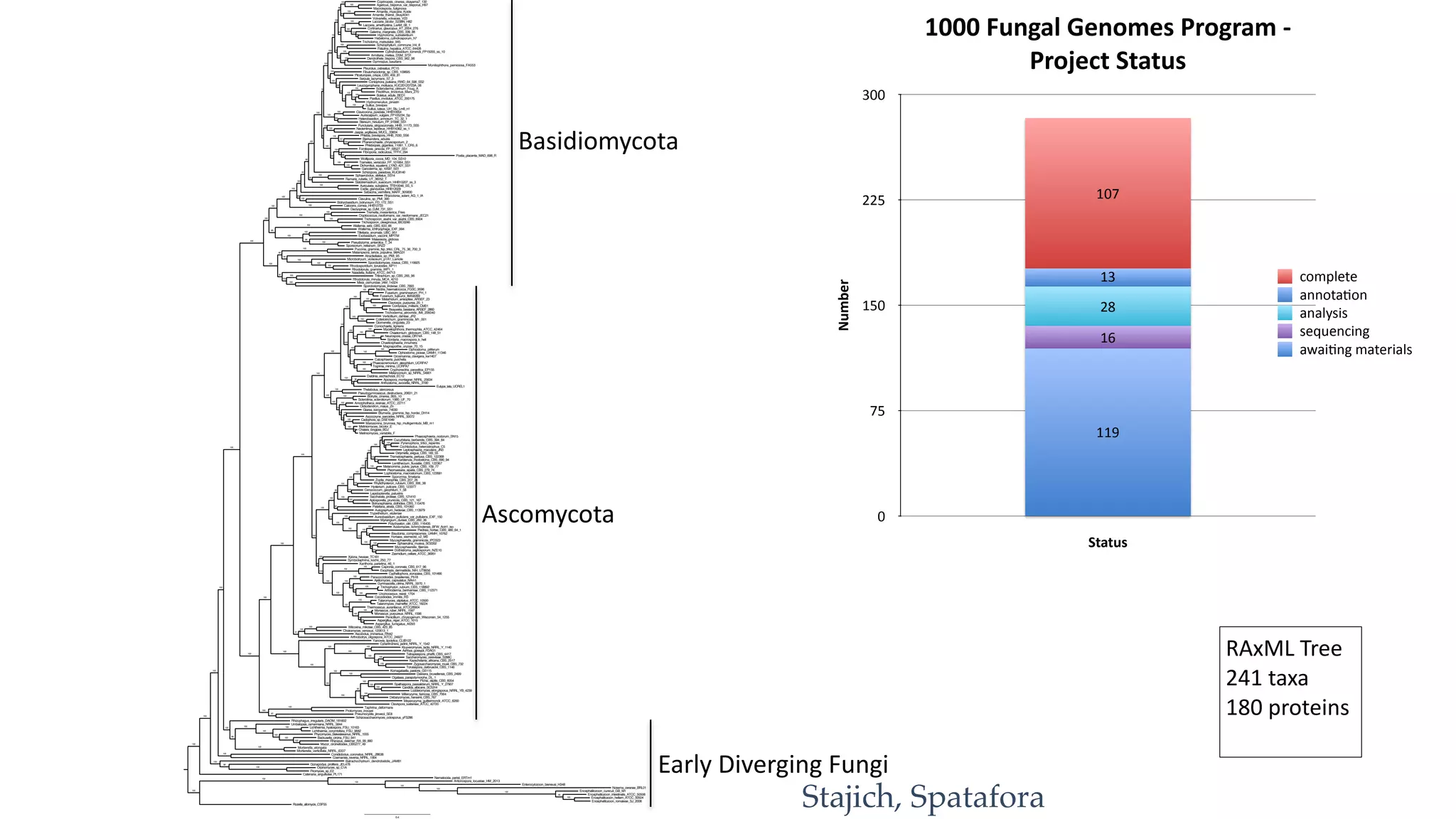
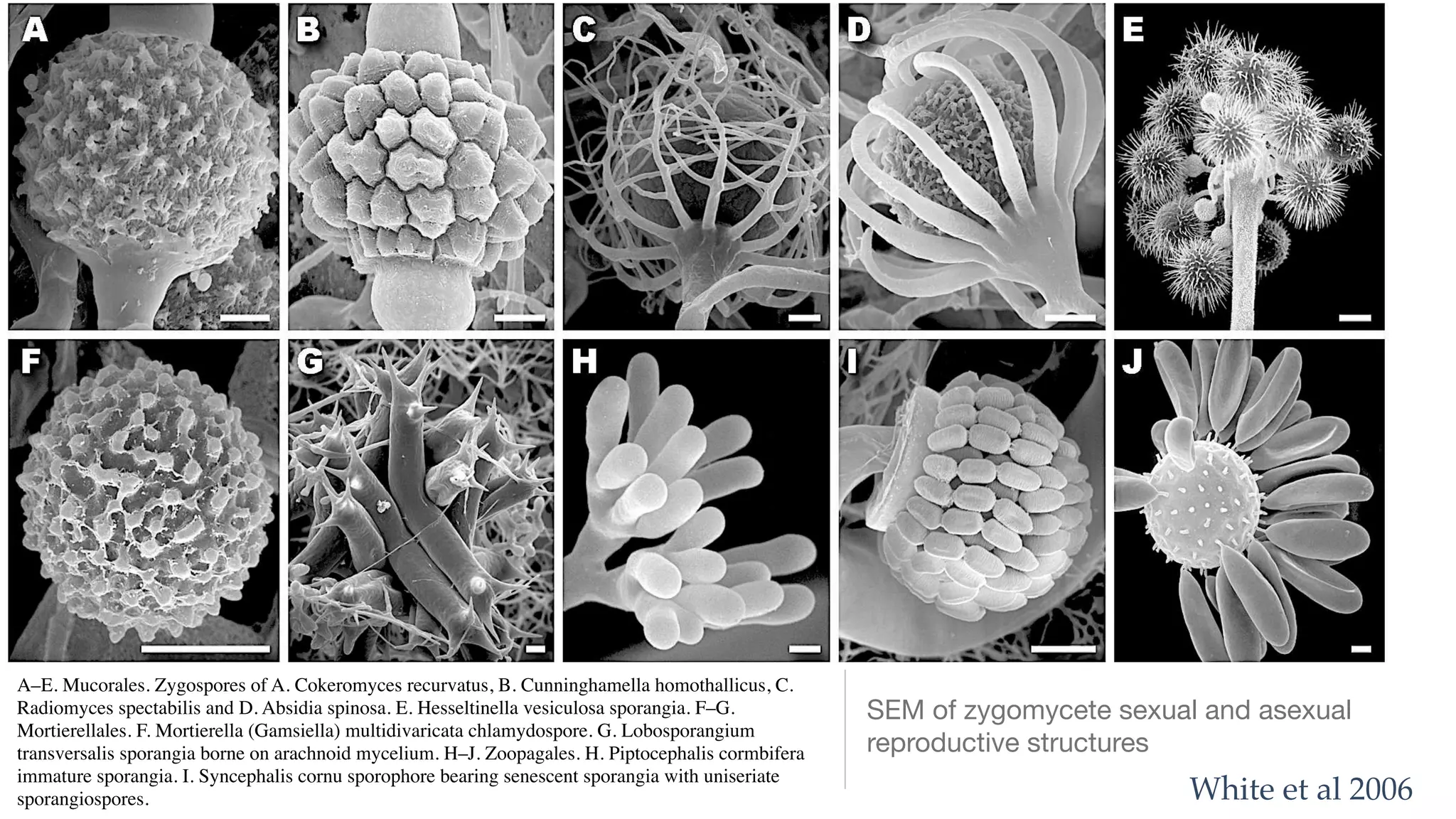
The document discusses exploring the phylogeny and genome evolution of zygomycete lineages through phylogenomics. The goals are to facilitate fungal genome sequencing across the kingdom fungi, produce a genome-scale phylogeny of fungi, create genome reference datasets, and obtain data from non-model organisms.