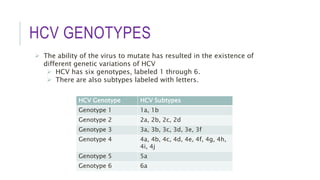
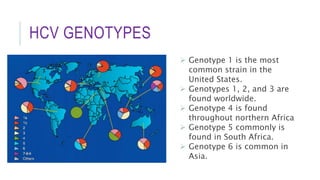
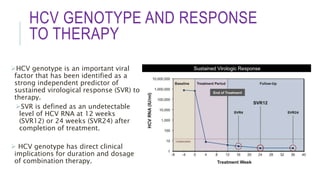
This document discusses hepatitis C virus (HCV) genotypes. It begins by introducing HCV and its effects, noting there are 6 major genotypes. Genotype 1 is most common in the US while genotypes 1, 2, and 3 are found worldwide. Genotype helps determine treatment response, as genotypes 1 and 3 are associated with more aggressive disease. The document then explores molecular genotyping as the "gold standard" method and alternative methods. It concludes by examining how HCV genotype predicts response to interferon-alpha therapy and newer direct-acting antiviral drugs, with genotypes 2 and 3 typically responding better to treatment.