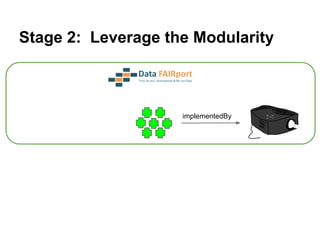
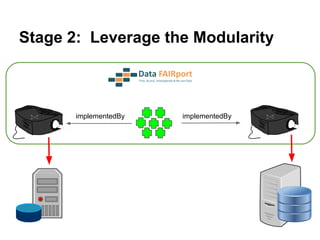
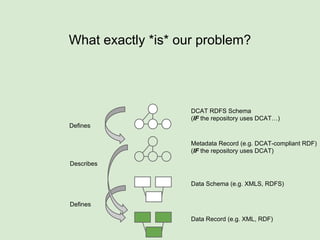
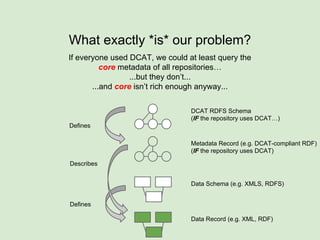
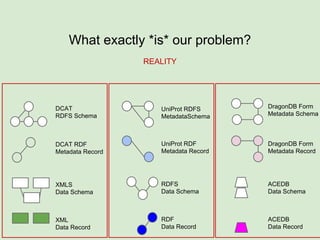
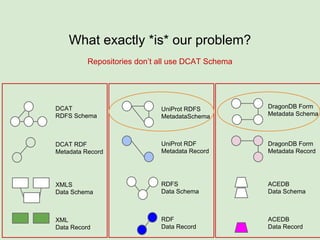
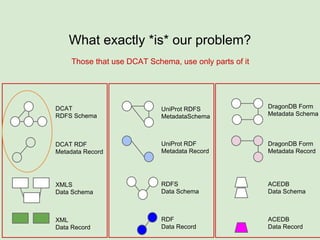
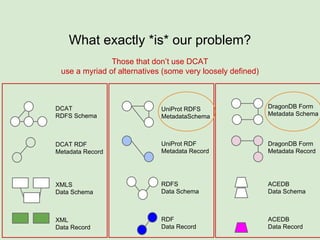
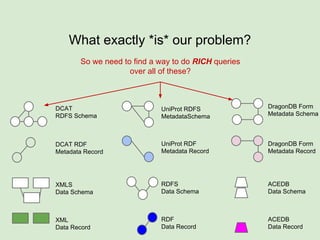
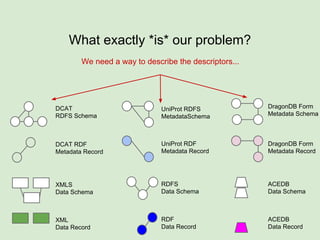
The document discusses the Fairport project aimed at improving the accessibility and interoperability of diverse data repositories. It addresses challenges such as the lack of standardized metadata structures and the need for unified access layers to enable seamless data discovery and integration. The project proposes solutions like shared metadata descriptors, known as fair profiles, and modular web services to facilitate effective data management and retrieval across varying data formats.


















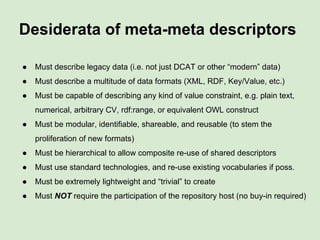

![Exemplar use-cases:
● A piece of software that can generate a “sensible”
data submission form for any repository
(at the Force 2015 meeting a few months ago I gave a presentation of a working
example of this… so I won’t repeat that today…)
● A piece of software that can generate a “sensible”
query form/interface for any repository
(demonstration of this today!)
Skunkworks Task #1 - [F]indable
Invent harmonized cross-repository meta-
descriptors](https://image.slidesharecdn.com/wilkinsondataversebostonworkshoppresentation-150618174924-lva1-app6892/85/Data-FAIRport-Skunkworks-Common-Repository-Access-Via-Meta-Metadata-Descriptors-by-Mark-Wilkinson-31-320.jpg)






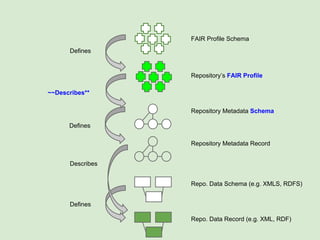












![Skunkworks Task #2 - [A]cessible
Are there already access layer definitions?](https://image.slidesharecdn.com/wilkinsondataversebostonworkshoppresentation-150618174924-lva1-app6892/85/Data-FAIRport-Skunkworks-Common-Repository-Access-Via-Meta-Metadata-Descriptors-by-Mark-Wilkinson-51-320.jpg)
![A set of behaviors for providing a unified (albeit simplistic!)
access layer for “records” contained in any Web resource
Skunkworks Task #2 - [A]cessible
Are there already access layer definitions?](https://image.slidesharecdn.com/wilkinsondataversebostonworkshoppresentation-150618174924-lva1-app6892/85/Data-FAIRport-Skunkworks-Common-Repository-Access-Via-Meta-Metadata-Descriptors-by-Mark-Wilkinson-52-320.jpg)









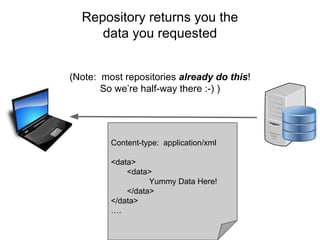


![Skunkworks Task #3 - [I]nteroperable
This is “the holy grail”!!](https://image.slidesharecdn.com/wilkinsondataversebostonworkshoppresentation-150618174924-lva1-app6892/85/Data-FAIRport-Skunkworks-Common-Repository-Access-Via-Meta-Metadata-Descriptors-by-Mark-Wilkinson-65-320.jpg)
![Skunkworks Task #3 - [I]nteroperable
This is “the holy grail”!!
This is where the FAIR Profile reveals its utility
“what it IS” vs. “what it IS NOT”](https://image.slidesharecdn.com/wilkinsondataversebostonworkshoppresentation-150618174924-lva1-app6892/85/Data-FAIRport-Skunkworks-Common-Repository-Access-Via-Meta-Metadata-Descriptors-by-Mark-Wilkinson-66-320.jpg)

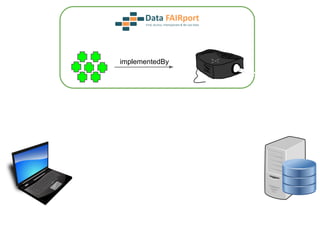
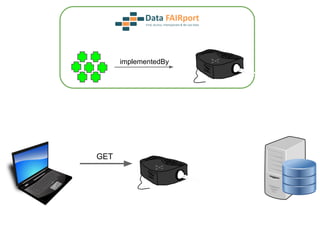
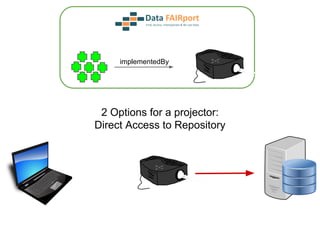
![Skunkworks Task #3 - [I]nteroperable
“FAIR Projectors”
A FAIR Projector is a (potentially) small, modular,
reusable Web based service that “projects” data
from a repository into the format
described by a FAIR Profile](https://image.slidesharecdn.com/wilkinsondataversebostonworkshoppresentation-150618174924-lva1-app6892/85/Data-FAIRport-Skunkworks-Common-Repository-Access-Via-Meta-Metadata-Descriptors-by-Mark-Wilkinson-68-320.jpg)
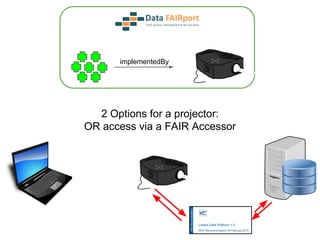
![Skunkworks Task #3 - [I]nteroperable
“FAIR Projectors”
A FAIR Projector is a (potentially) small, modular,
reusable Web based service that “projects” data
from a repository into the format
described by a FAIR Profile
http://linkeddatafragments.org/](https://image.slidesharecdn.com/wilkinsondataversebostonworkshoppresentation-150618174924-lva1-app6892/85/Data-FAIRport-Skunkworks-Common-Repository-Access-Via-Meta-Metadata-Descriptors-by-Mark-Wilkinson-69-320.jpg)