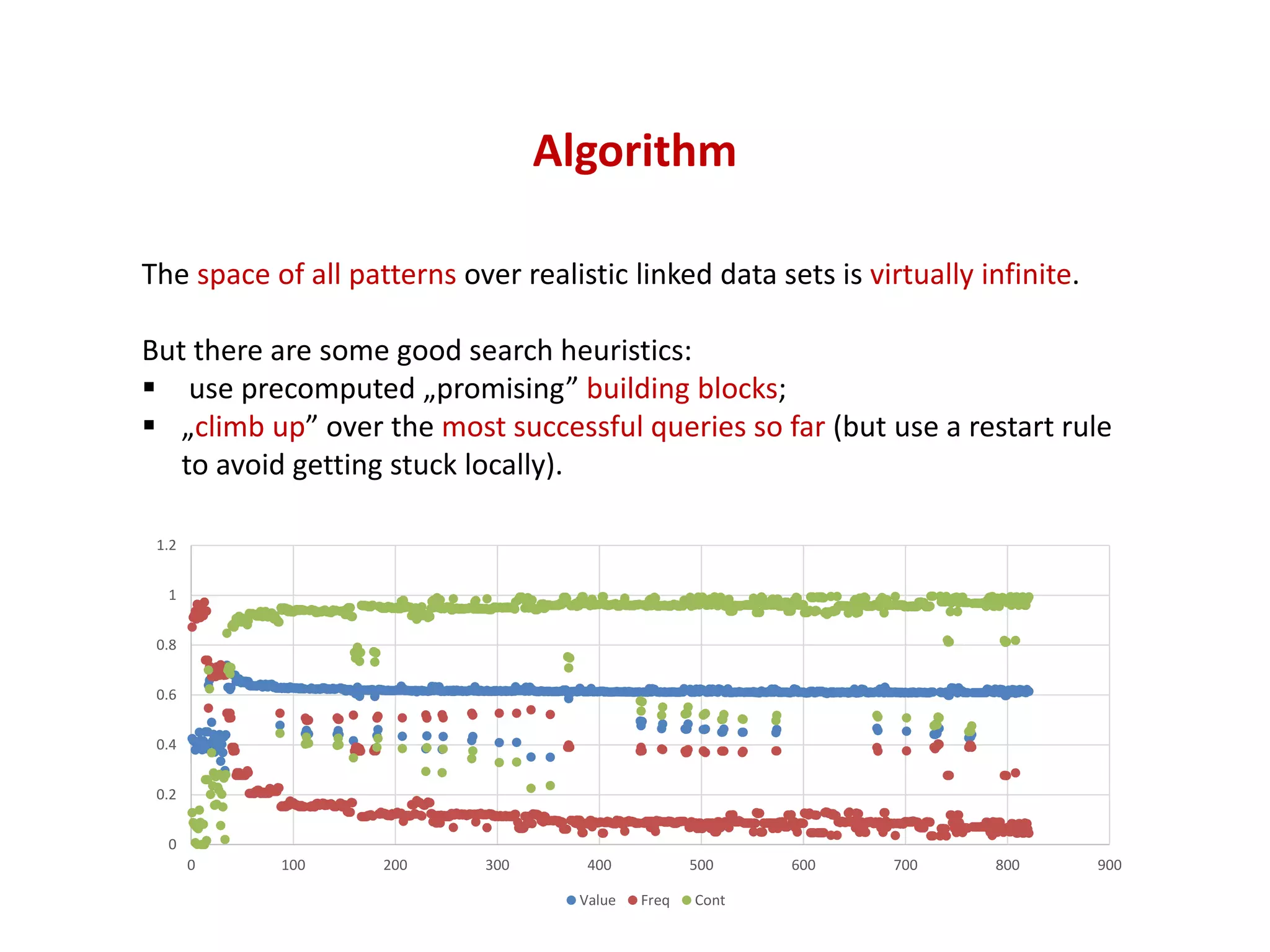
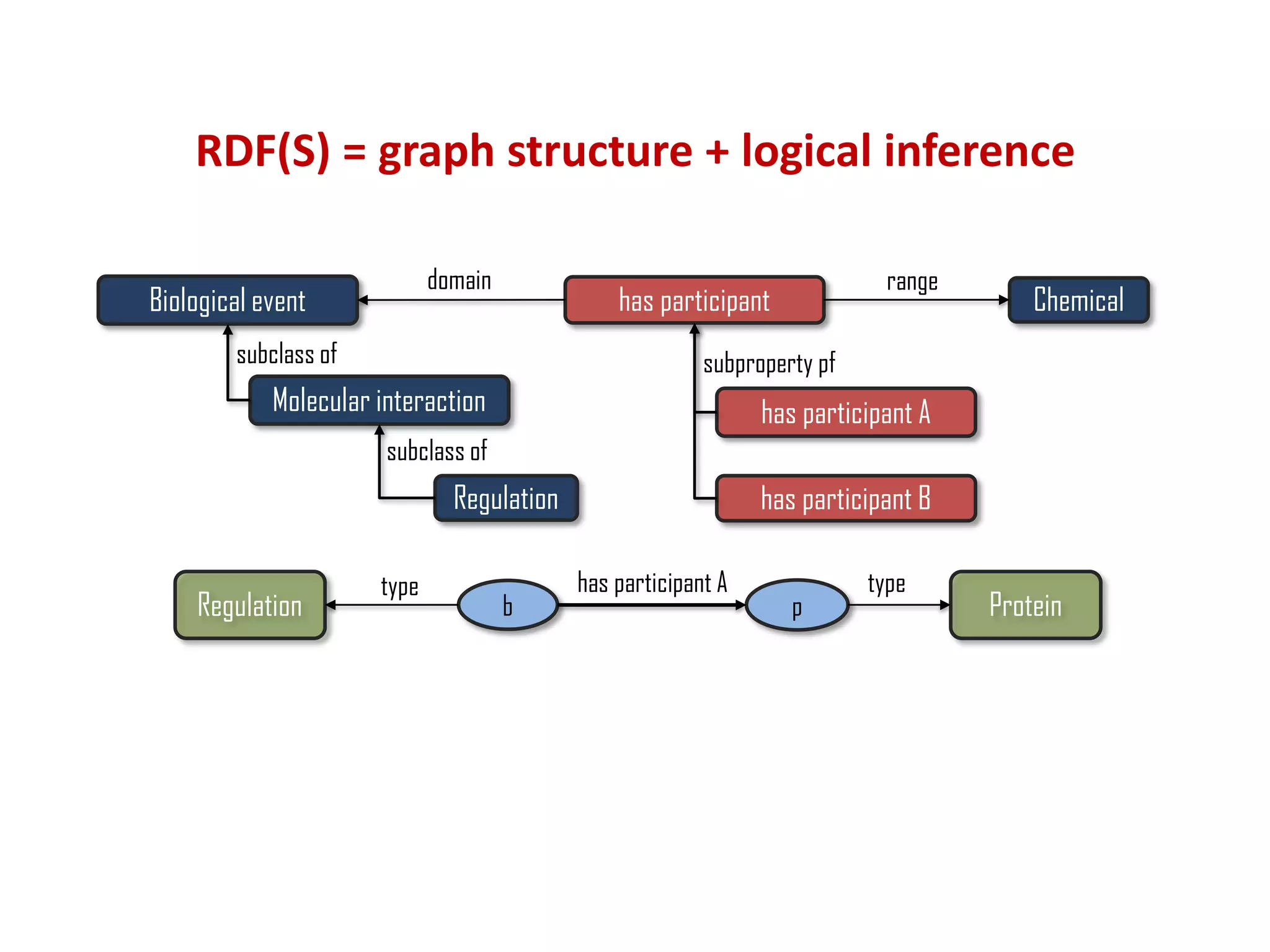
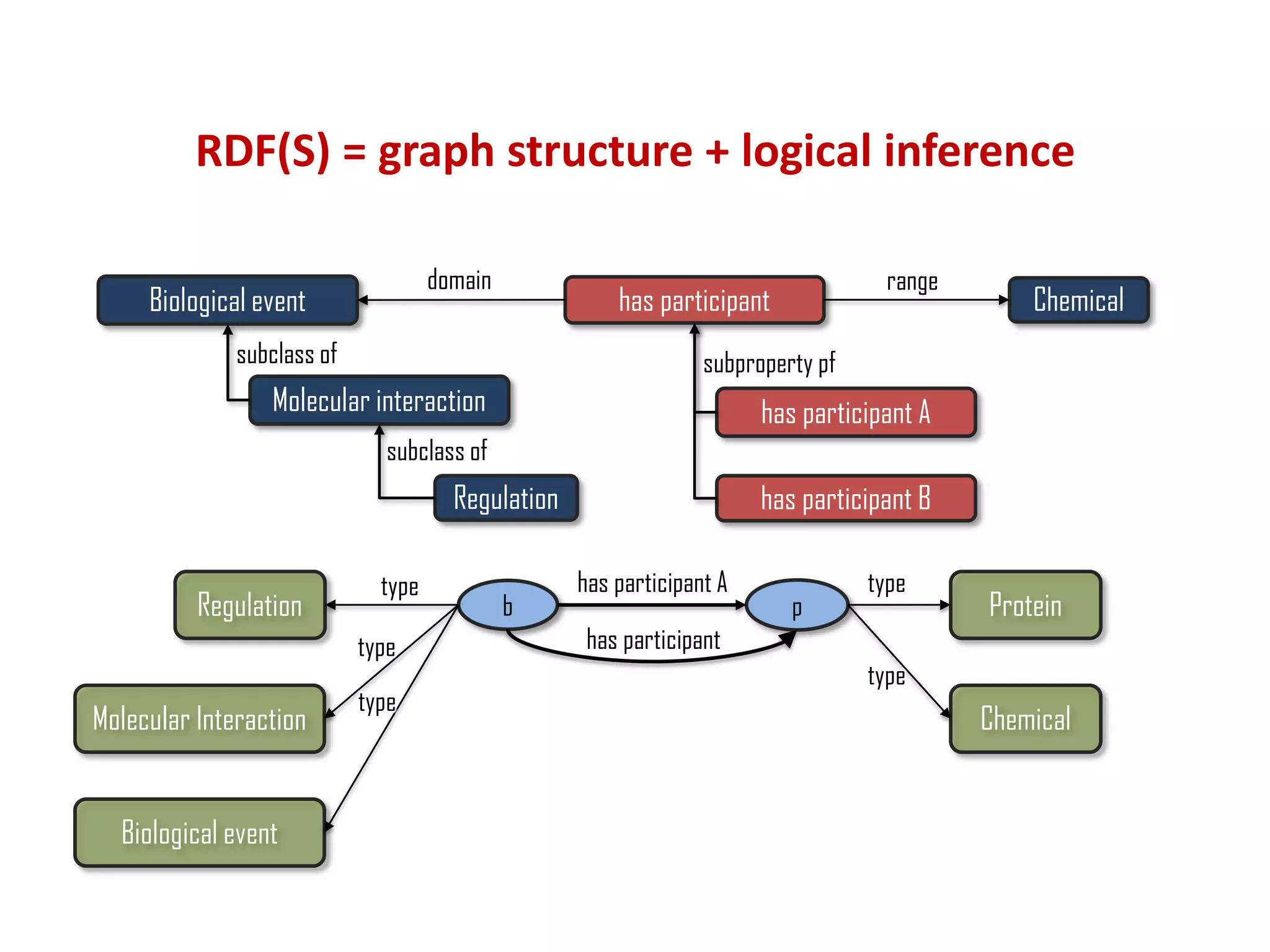
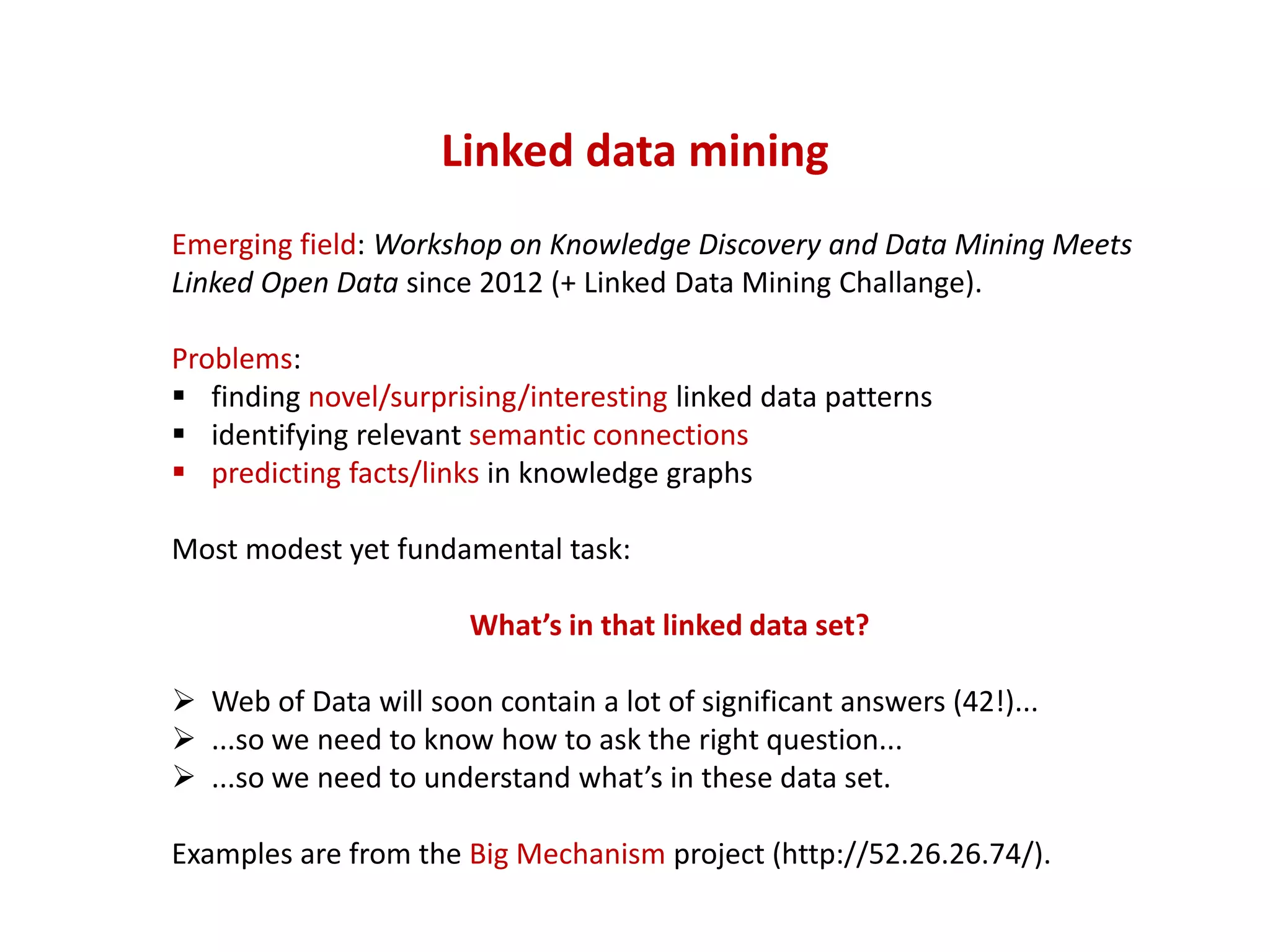
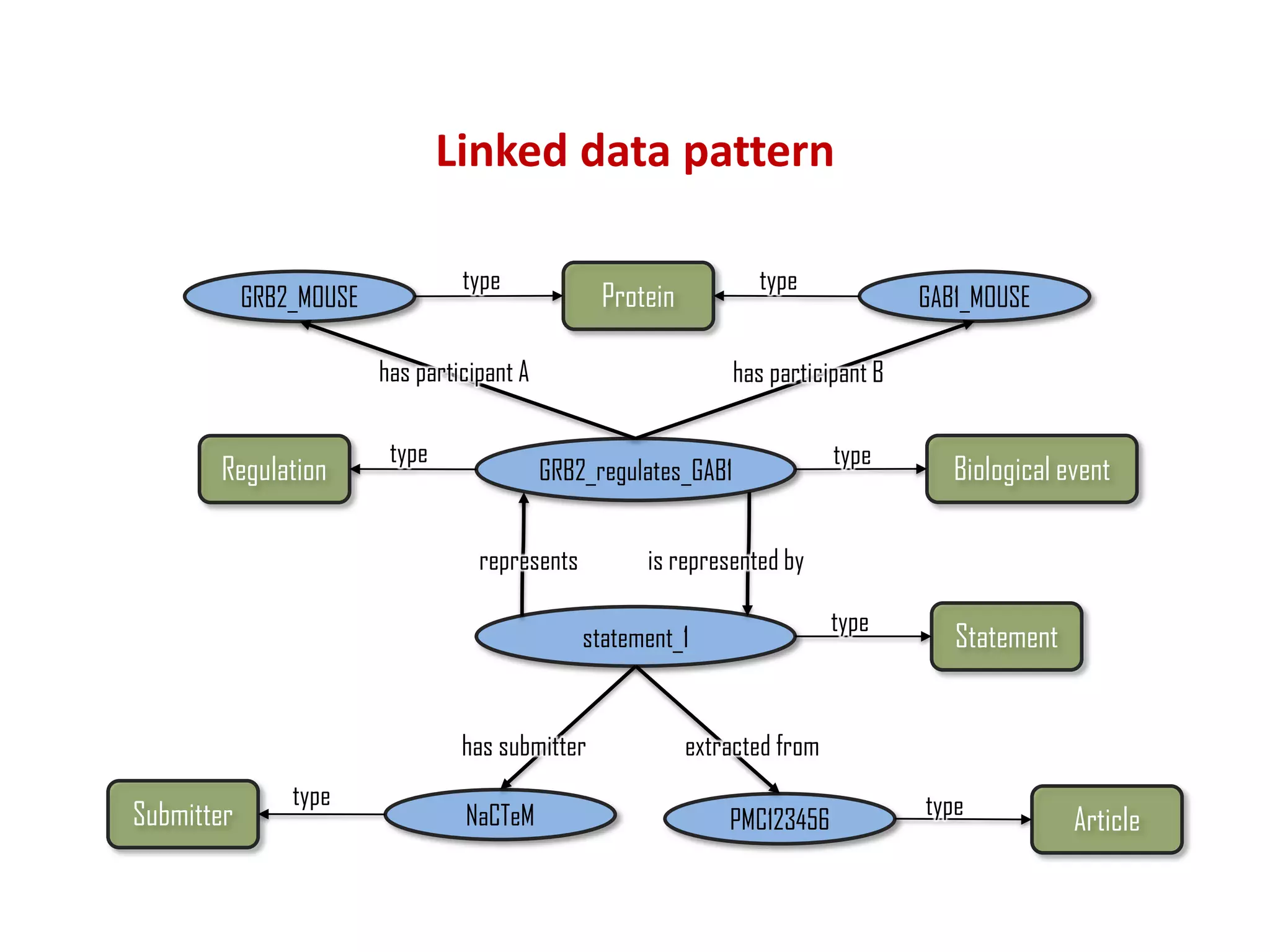
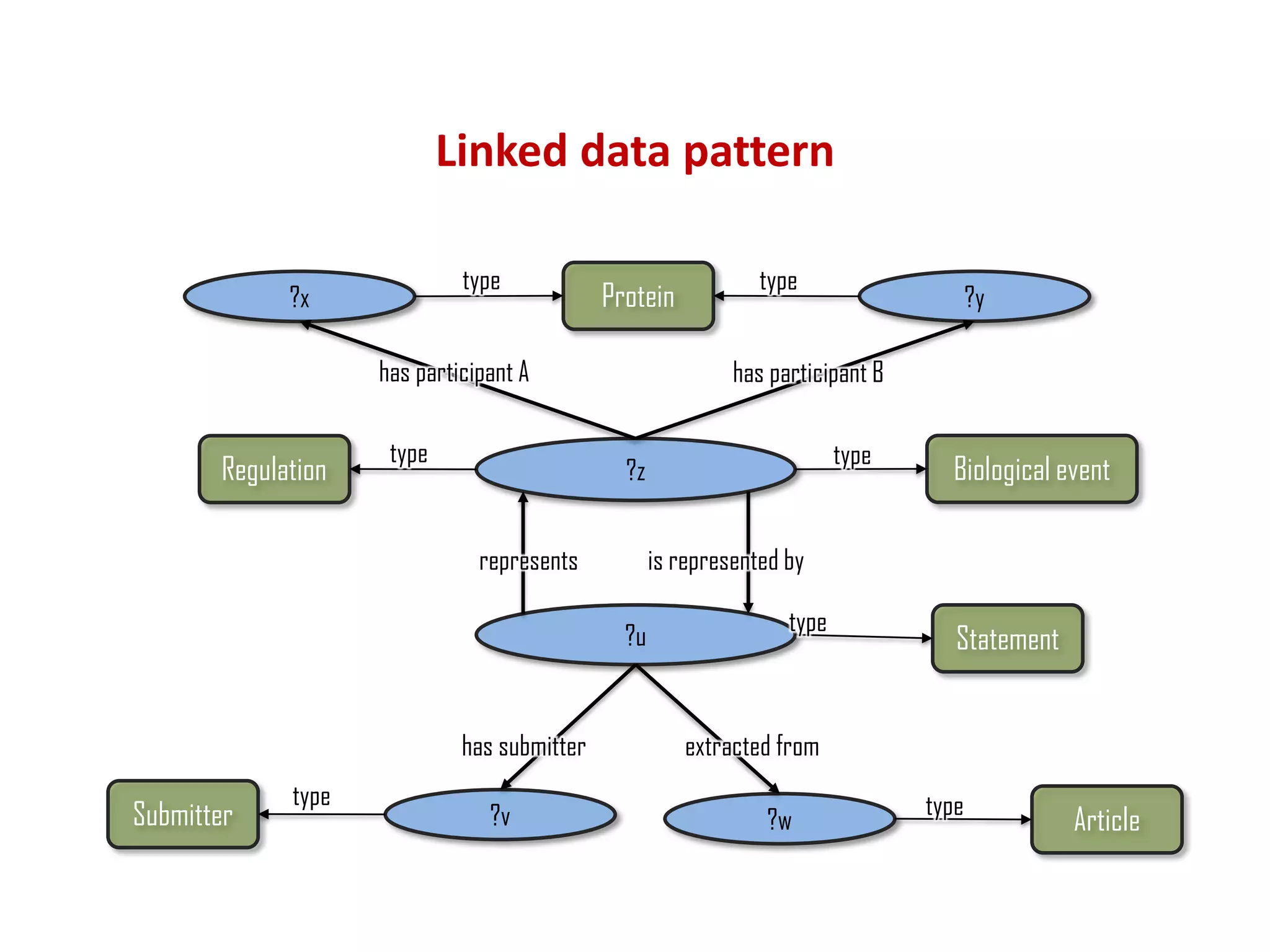
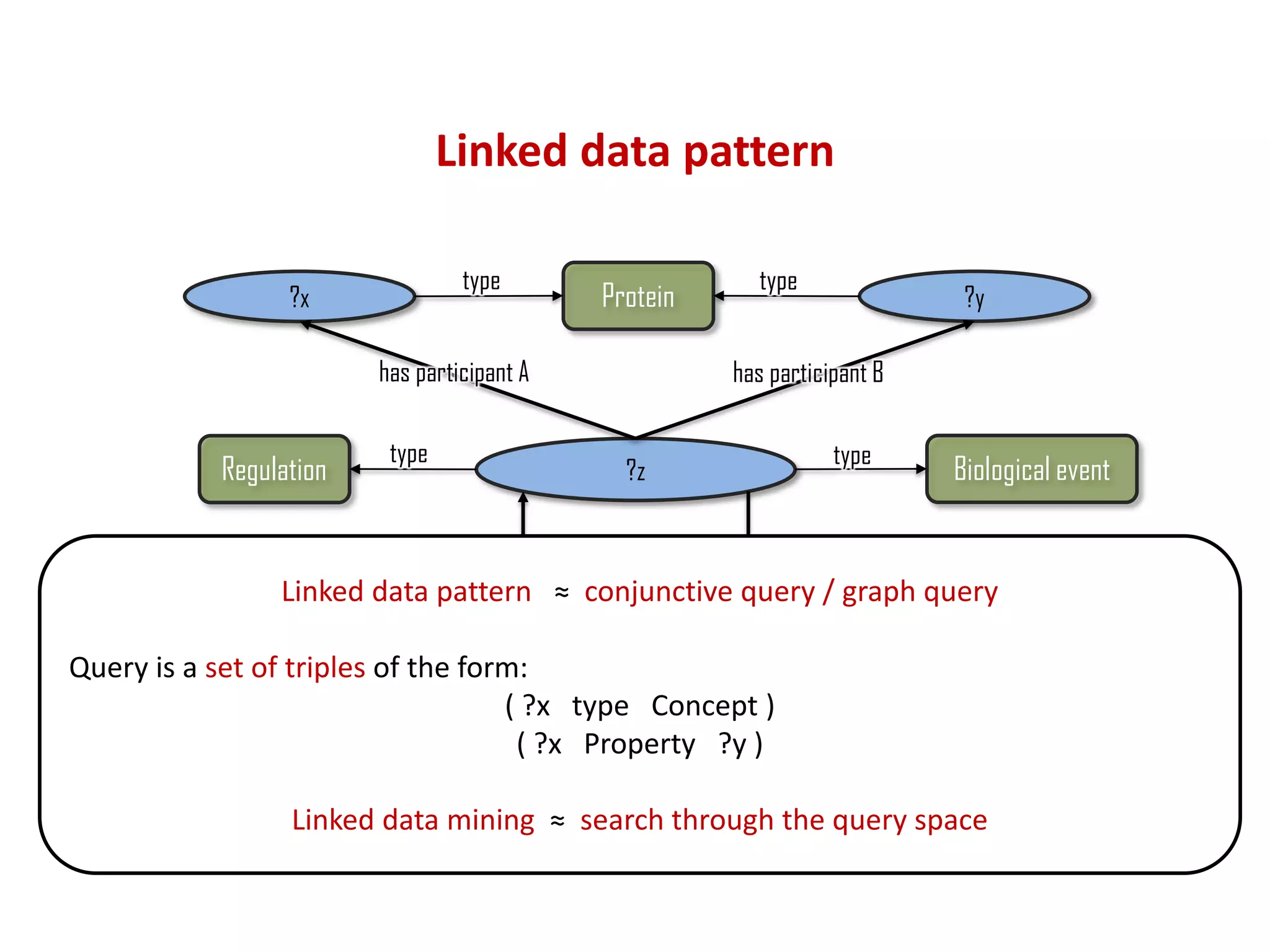
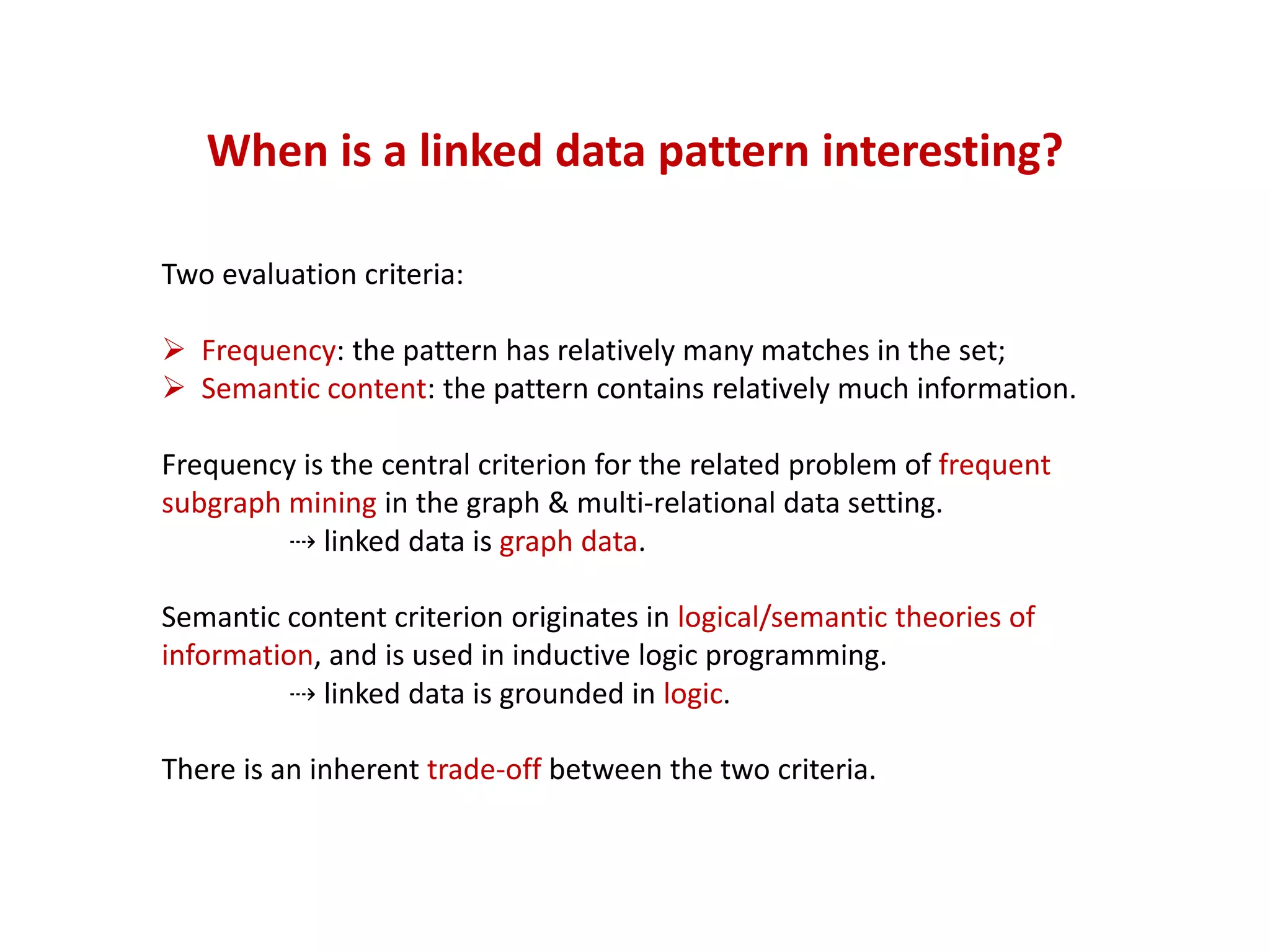
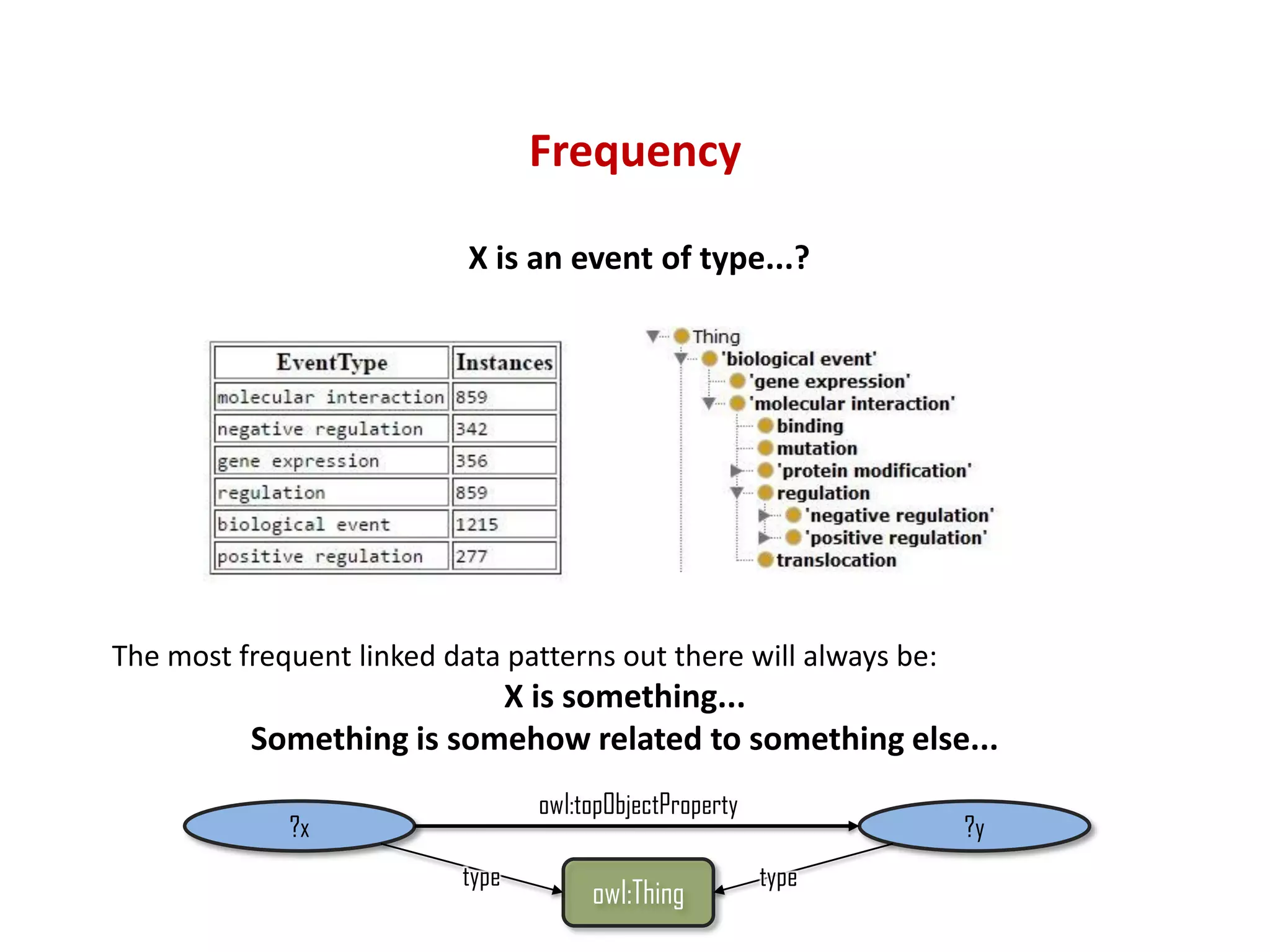
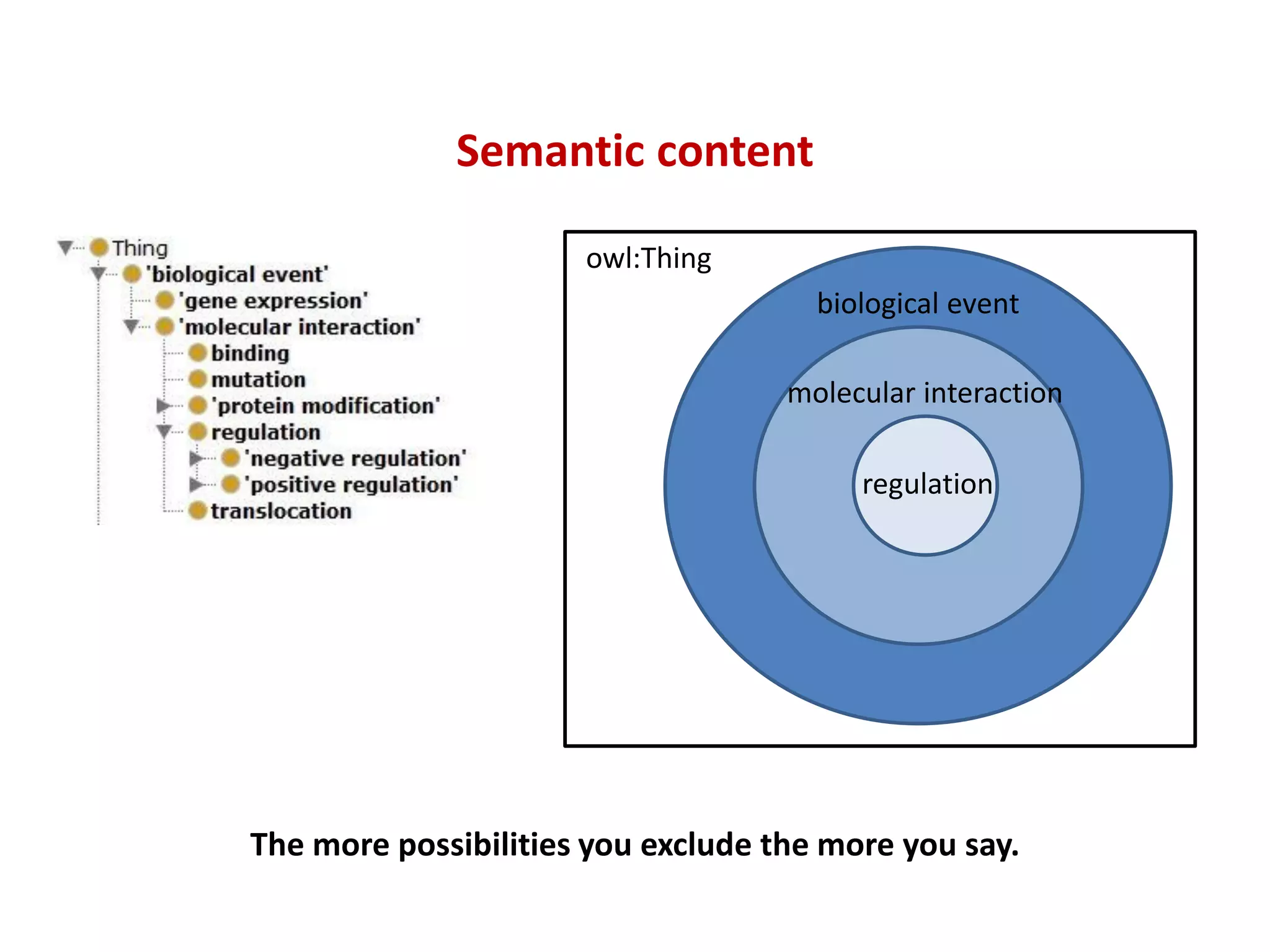
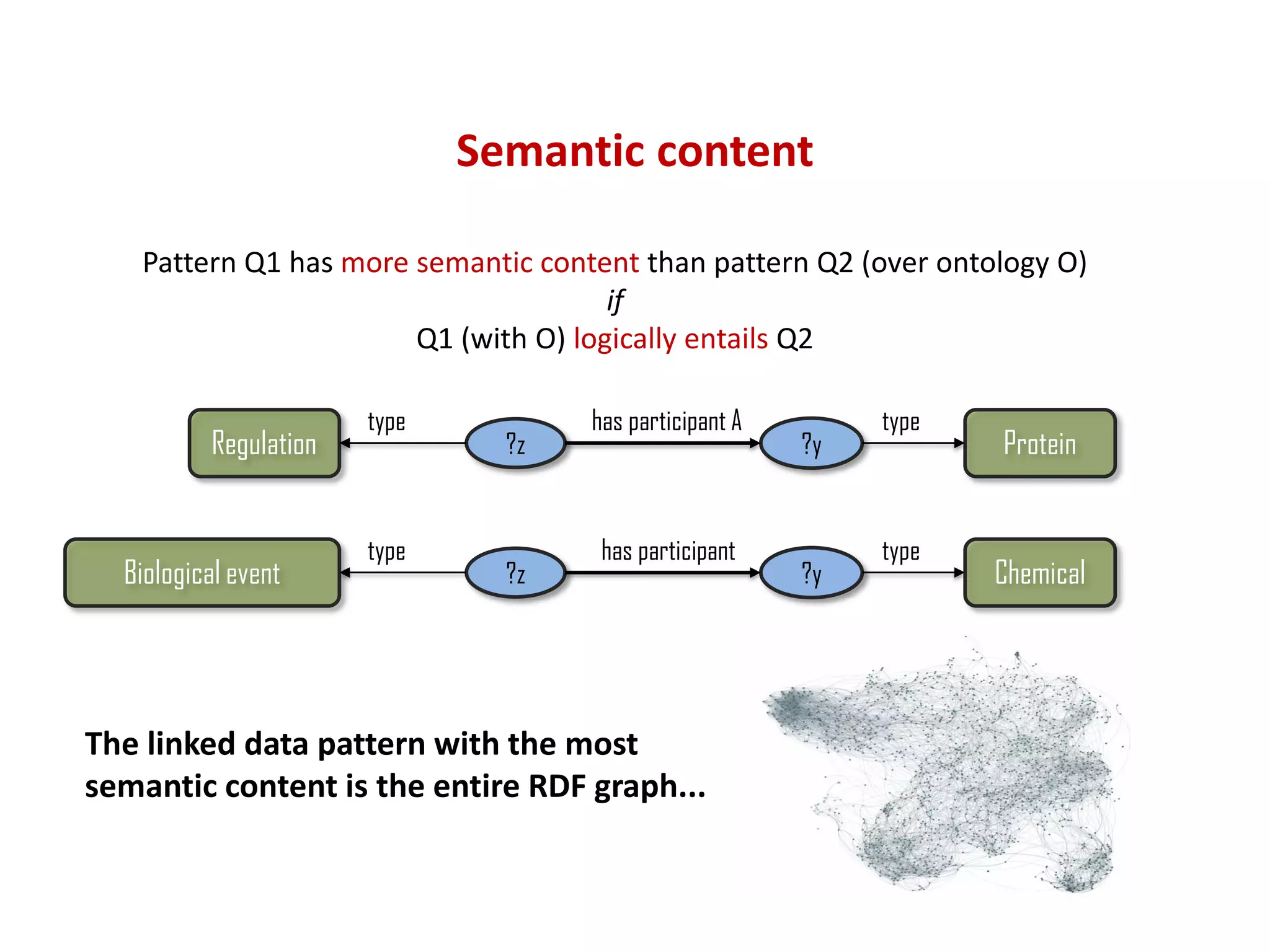
The document discusses the importance of linked data patterns, highlighting their flexibility, machine accessibility, and potential for knowledge discovery. It emphasizes two evaluation criteria for interesting patterns: frequency of matches and semantic content, while addressing the challenges and methodologies in linked data mining. The author suggests a need for novel approaches and principles in the field to enhance understanding of linked datasets.




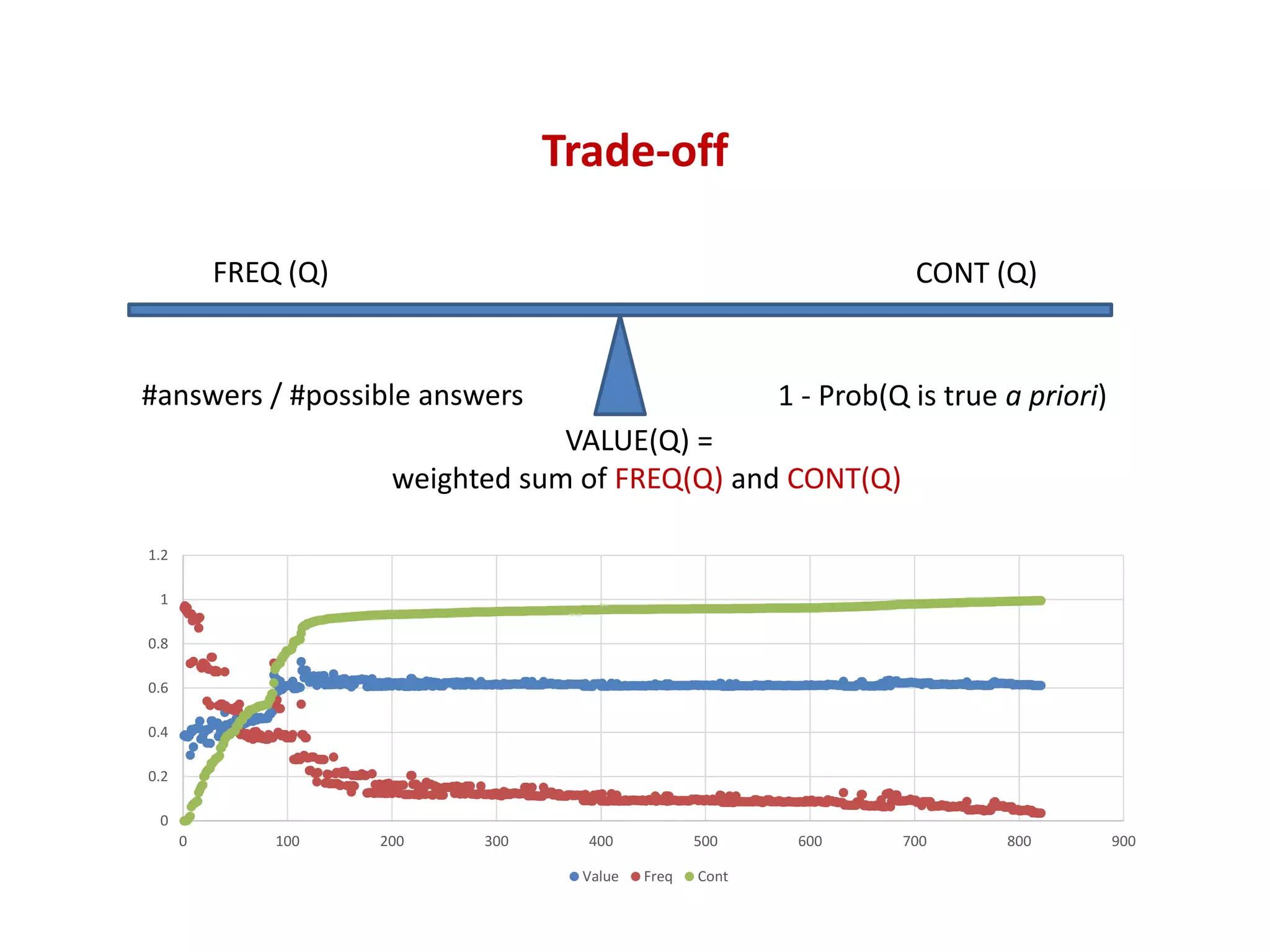
![Trade-off
0
0.2
0.4
0.6
0.8
1
1.2
0 100 200 300 400 500 600 700 800 900
Value Freq Cont
Q1 = textual_entity(x)
Q2 = statement(x)
Q3 = event(x)
Q4 = journal_article(x), published_in(x, u), journal(u),
is_extracted_from(w, x), statement(w), contained_in(w, y),
table(y), represents(w, v), negative_regulation(v),
has_submitter(y, z), submitter(z), [...] (10 variables)
Q5 = table(x), has_submitter(x, z), submitter(z), contains_statement(x, y), statement(y), contained_in(y, x)
Q6 = positive_regulation(z), is_represented_by(z, y), statement(y), represents(y, z), contained_in(y, x),
table(x), has_submitter(x, v), submitter(v), contains_statement(x, y).](https://image.slidesharecdn.com/klarman-meetup-2016-170419195300/75/What-makes-a-linked-data-pattern-interesting-24-2048.jpg)