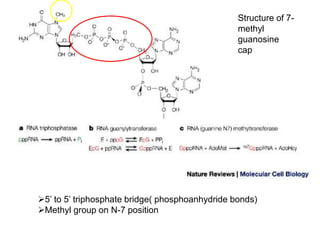
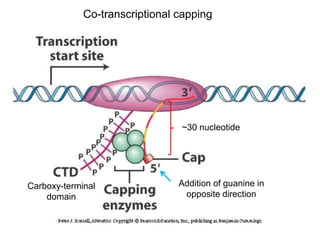
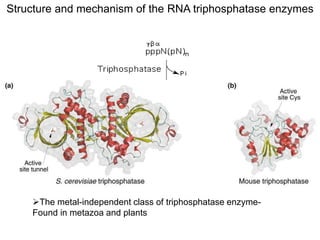
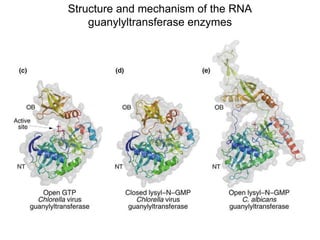
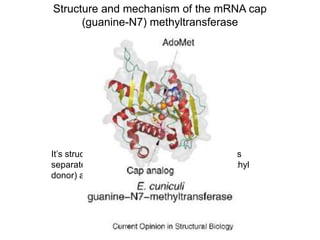
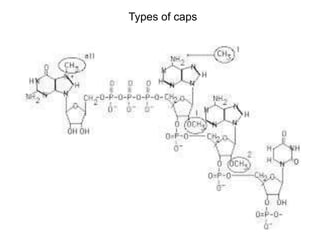
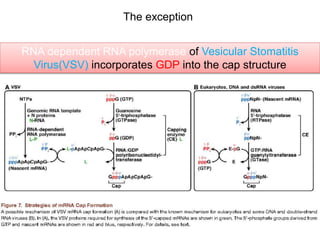
This document discusses the 5' terminal m7G cap structure found on eukaryotic messenger RNA. It summarizes that the cap evolved in eukaryotes to serve as an alternative to the prokaryotic Shine-Dalgarno sequence for directing ribosomes to mRNA. The structure of the cap involves a 5' to 5' triphosphate bridge and methyl group on the guanine nitrogen. Capping occurs co-transcriptionally within 30 nucleotides of the 5' end. The cap protects mRNA from degradation, marks the translation start site, and facilitates translation initiation and splicing.