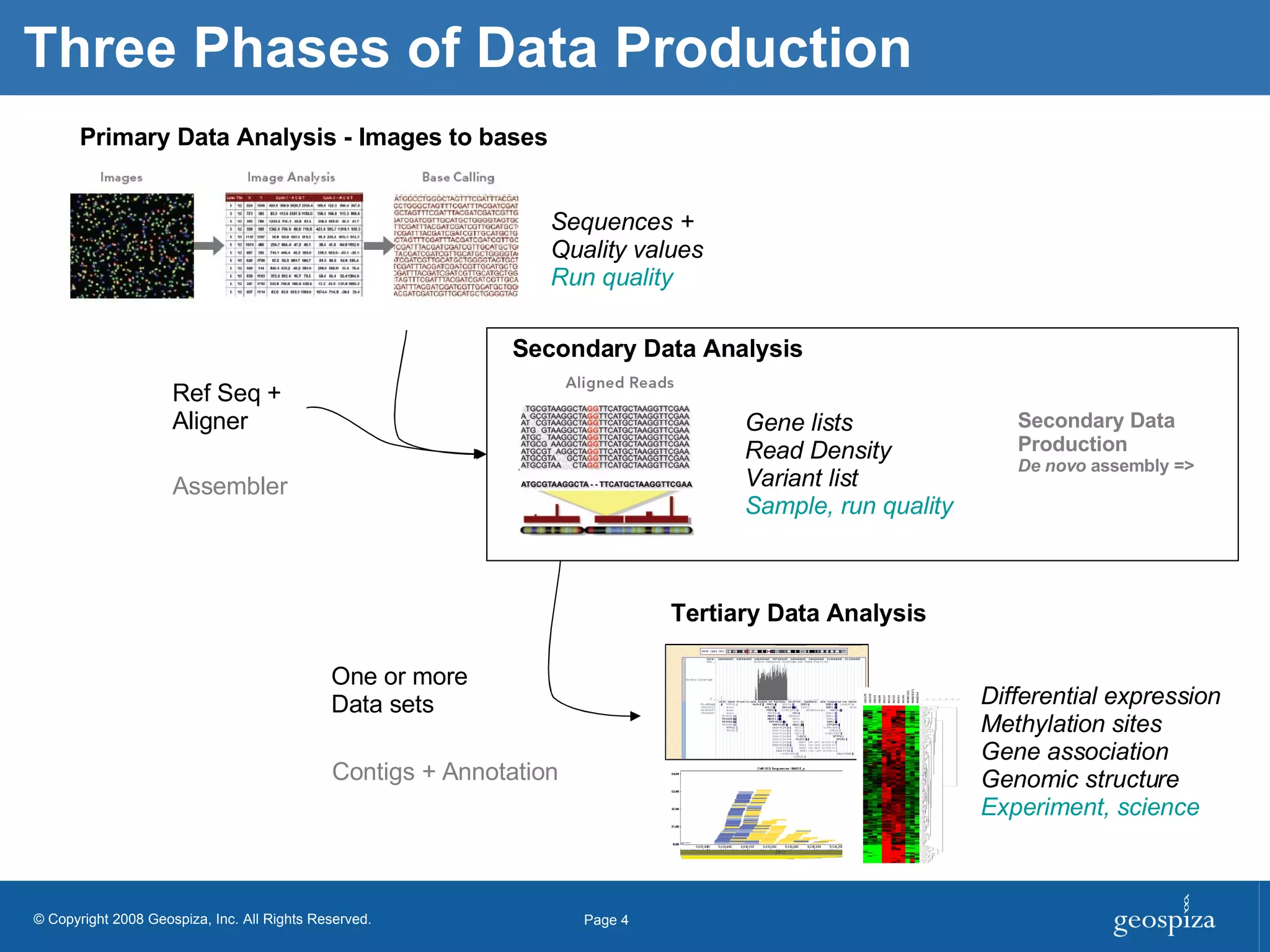
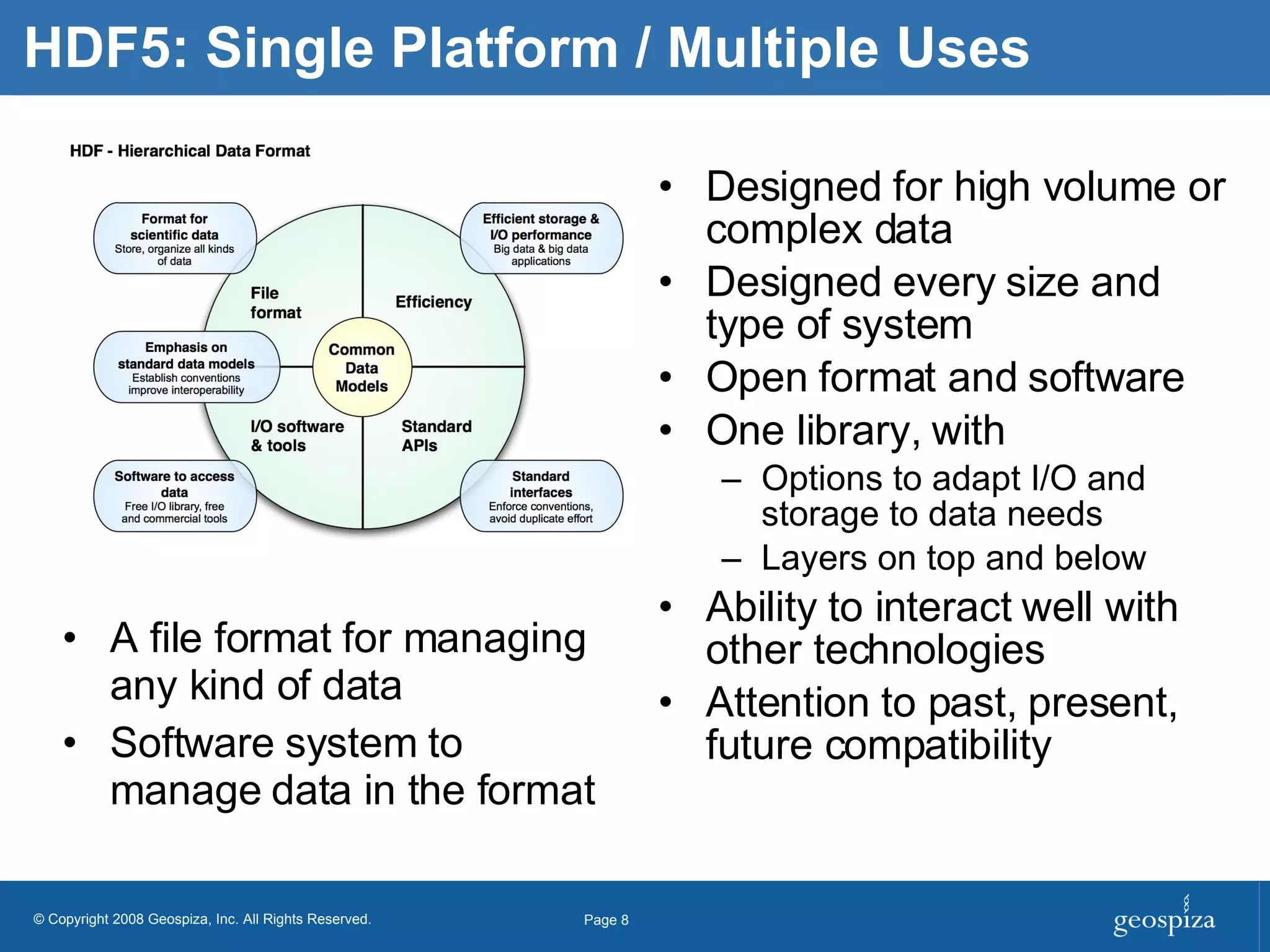
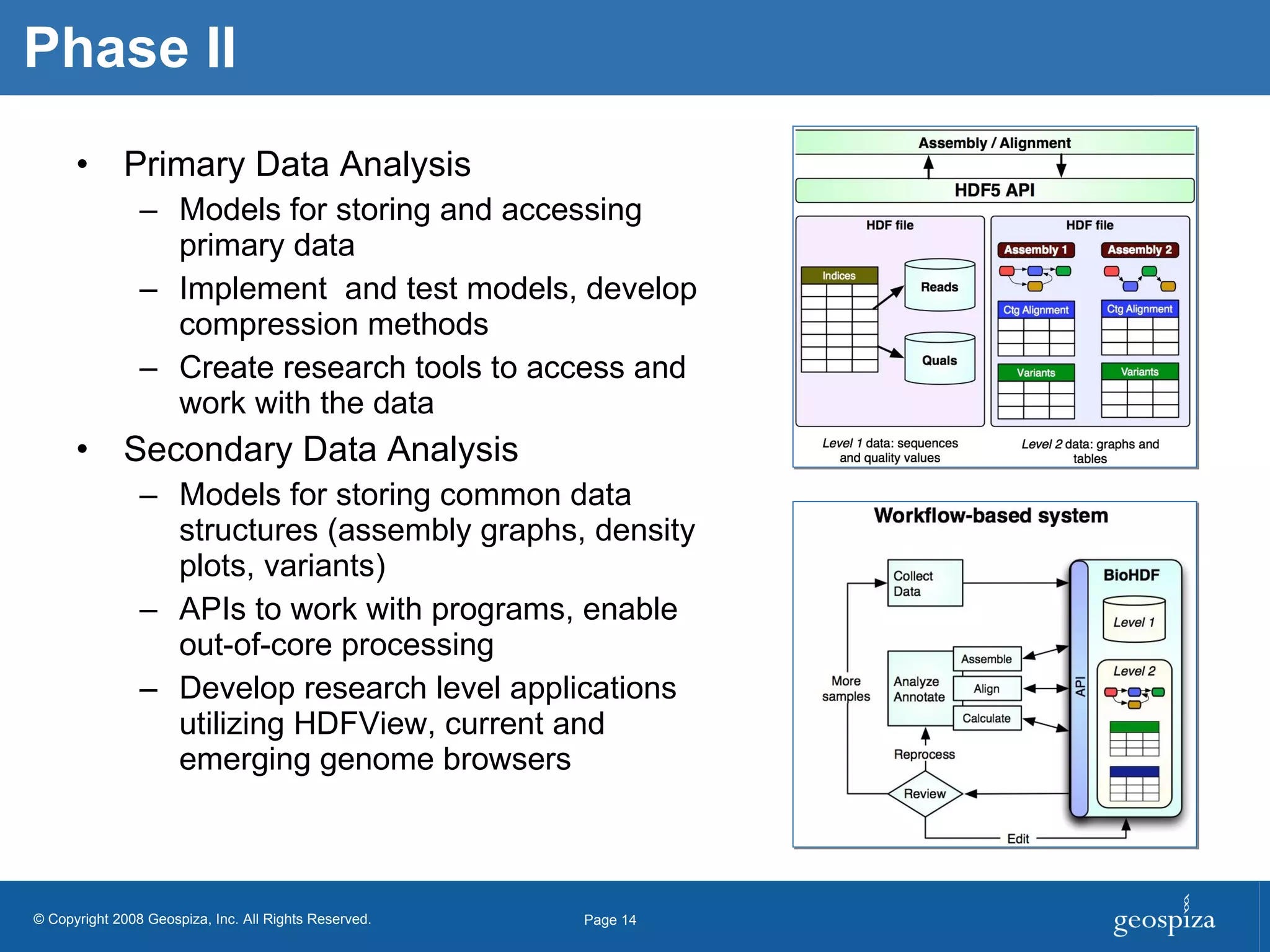
BioHDF is a project to develop open binary file formats and software tools for managing large-scale genomic data from next-generation DNA sequencing. The project aims to address challenges related to the proliferation of file formats, redundancy of data, and computational overhead by building on the HDF5 data model and libraries. BioHDF will develop models and applications to support primary and secondary data analysis from sequencing, with collaborations planned with software developers and research groups.

![Next Generation DNA Sequencing Next Gen Sequencing platforms produce ~1500 X more data than CE (Sanger) A single Next Gen instrument can produce 20 times more data a single run than a day’s operation of a genome center with 100 CE instruments In Sequence quotes - July 2007 Toby Bloom, Broad Institute “Next-gen sequencing i mpacts all aspects of informatics.” Phil Butcher, Sanger “ T he best way to move terabytes of data is still disk.” Want to process data closer to the machine. Eugen Clark, Harvard “[community] needs to start talks about data retention.” Kelly Carpenter, Wash U “these sequencers are going to totally screw you.” Nature Methods July 2008: “Byte-ing off more than you can chew”](https://image.slidesharecdn.com/smithtbiohdfbosc2008-1217538108837374-9/75/Smith-T-Bio-Hdf-Bosc2008-3-2048.jpg)