IB Chemistry on ICT, 3D software, Chimera, Jmol, Swiss PDB, Pymol for Internal Assessment on Hemoglobin, Myoglobin and Cytochrome
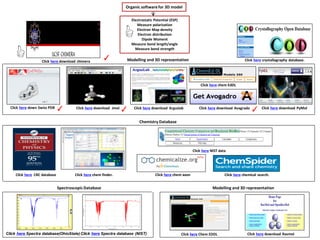
- 1. Electrostatic Potential (ESP) Measure polarization Electron Map density Electron distribution Dipole Moment Measure bond length/angle Measure bond strength Organic softwarefor 3D model Click here download Rasmol Click here download PyMolClick here download Jmol Click here Chem EDDL Click here chemical search.Click here CRC database Modelling and 3D representation Chemistry Database Click here Spectra database(OhioState) Click here Spectra database (NIST) Click here chem finder. Spectroscopic Database Click here down Swiss PDB Modelling and 3D representation Click here crystallography database. ✓ ✓ Click here NIST data ✓Click here download Arguslab Click here chem axon Click here download Avagrado Click here chem EdDL Click here download chimera ✓
- 2. Measure polarization Electron Map density Electron distribution Electrostatic Potential (ESP) Dipole Moment Measure bond length/angle Measure bond strength Organic softwarefor 3D model Click here download Rasmol Click here download PyMolClick here download Jmol Click here Chem EDDL Click here chemical search. Modelling and 3D representation Quick Chemistry DatabaseCheck Click here down Swiss PDB Modelling and 3D representation ✓ ✓ Click here NIST data ✓Click here download Arguslab Click here chemaxon quick chem check Click here download Avagrado Click here chem EdDL Click here for Visualization/3D sources Click here download Marvin Sketch Click here quick chemical check Click here quick chemical check
- 3. Measure bond length/angle Measure number H2 bonds Measure bond strength Protein 1, 2 , 3O structure Presence of disulfide bond Presence alpha and beta pleated sheet Protein Data Bank Protein database key in - PDB 4HHB Click here Chimera tutorial 1 2 Uses molecular modelling 1 2 Chemicalviewer 3D structure(Chimera) Download PDB text file File – fetch by ID- 4HHB Select – residue – HEM Select – chain A – Action – Ribbon – Hide Select – chain B,C,D - Action – ribbon Hide Display only ligand Heme Tool- structural analysis - Distance Select 2 atom -by press control/shift/left click select 2 points Tool – structure analysis – create to get distance 3 Check here 4HHB Chimera 1MBO Select Histidine that are close to ring Locate His F8 and E7 Make measurement Click here download chimera Tool – Sequence – choose sequence for 4 chains Identify amino acids of interest 4
- 4. Type PDB code – 4HHB Right click – select Hetero Select - HETATM – HEM 4 Heme is display from 4 chains Measure bond length/angle Measure number H2 bonds Measure bond strength Protein 1, 2 , 3O structure Presence of disulfide bond Presence alpha and beta pleated sheet Click here J mol protein video Chemical viewer 3D structure (Jmol) Uses molecular modelling 1 J mol executable file Measure distance Select measure – distance for porphyrin ring Measure ring size/distance Fe from plane Select protein – by residue – Histidine Measure and locate His F8 Measure and locate His E7 final heme – click here J mol executable file 1 Type 4HHB into protein data bank Look for ligand Heme Model kit to design molecule Click here deoxyhemoglobin chimera 2 2 3 4 3 4 final product All histidine shown Get structure from PDB and MOL
- 5. Measure bond length/angle Measure number H2 bonds Measure bond strength Protein 1, 2 , 3O structure Presence of disulfide bond Presence alpha and beta pleated sheet Organic softwarefor 3D model (Pymol) 1 1 Click here - Protein Data Bank Protein database key in - PDB 4 letter code 3 Click here download PyMol Click here Pymol video tutorial Click file – open your download pdb file from Protein Data bank Get to command term – Type fetch 4HHB H - Hide – S - Show cartoon – C – Type by ss Select 4 Hem – Look for 4 Hemes Select 4HHB – H – hide everything Select Heme – Show stick Look His – select and name His F8 and His E7 2 Press S – sequence at bottom screen. Right click – zoom in Select HEM - hemoglobin 4 Uses molecular modelling 2 Select heme – right click – action – around 5A Look for His F8 and E7 around heme Make measure for distance Double click to display name of atom
- 6. Measure bond length/angle Measure number H2 bonds Measure bond strength Protein 1, 2 , 3O structure Presence of disulfide bond Presence alpha and beta pleated sheet Protein Data Bank Protein database key in - PDB 4HHB Click here Swiss PDB tutorial 1 2 Uses molecular modelling 1 2 Chemicalviewer 3D structure(Swiss PDB) Download PDB text file File – open 4HHB pdb downloaded from databank Window – Control panel Remove – side chain Select – Group kind – HETATM Display – stereo view Show only selected 4 Heme Click here down Swiss PDB Select – Group kind – Histidine Select – Residue – close to 2A Locate Histidine and make measurement 3 Check for heme and Histidine only from control panel Select Histidine that are close to ring Locate His F8 and E7 and make measurement 4
- 7. Heme Hemoglobin - 4 chain - 4 heme - 4 Fe 2+ Fe2+ Heme (porphyrin) Hemoglobin - 2 alpha chain - 144 amino acid - 2 beta chain - 146 amino acid Myoglobin - 1 chain – 1 heme – 1 Fe2+ - 154 amino acids HemoglobinMyoglobin Fe2+ Heme (porphyrin) PDB code files Oxyhemoglobin – 1GZX, 1JY7 Deoxyhemoglobin – 1A3N, 4HHB, 2HHB, 1HBB, 1G9V, 101J Myoglobin – 4MBN, 3RGK, 5MBN Fetal hemoglobin – 1FDM Sickle cell Hemoglobin - 2HBS, 1NEJ Cytochrome(bovine) - 2B4Z Cytochrome(horse) - 1HRC Cytochrome(yeast) - 1YCC Cytochrome(human) - 3NWV PDB file type for data analysis 1 Analyze using Chimera/Pymol/Swiss PDB/Jmol 1 Chimera Swiss PDB JmolPymol Fe in cytochrome 1 polypeptide chain Fe2+ Cytochrome c
- 8. Possible ResearchQuestion Measuring using 3D modelling Data Collection using 3D modelling Data Collection using Database Click here Jmol Click here PyMol - What is the distance bet Fe and His E7 and F8, and are they the same for diff heme found in hemoglobin/myoglobin/Cytochrome - Is His E7/F8 orientation similar for Hemoglobin, Myoglobin, Cytochrome - Is there any differencesbet distance/position/orientationof porphyrin ring for Hemoglobin, Myoglobin, Cytochrome - How is Fe2+located, along or out of plane for Hemo/Myoglobin/Cytochrome - Is distancebet Fe and ligand N of porphyrin the same for Hemoglobin/Myoglobin/Cytochrome - Is there any variation in terms of Fe and His E7/F8 for Hemoglobin/Myoglobin/Cytochrome - Why His E7 and F8 are locatedin such a way across many different species? Is their orientation highly conserved and why? - Similarity among cytochromes found in diff species of organism Click here NCBI Click here UCSC Click here Ensembl Structuralsimilarity and diff bet Hemoglobin/Myoglobin and Cytochrome Myoglobin Hemoglobin Cytochrome Hemoglobin Chimera Pymol Jmol Swiss PDB Mean Orientation His/Fe Similar Similar Similar Similar Similar Bond length N - Fe 2.12A 1.90A 2.02A 2.02A 2.01A Bond length Fe – E7 5.93A 5.80A 5.45A 5.42A 5.55A Bond length Fe – F8 2.25A 2.05A 2.10A 2.21A 2.13A Chimera Swiss PDB Data source Myoglobin Chimera Pymol Jmol Swiss PDB Mean Orientation His/Fe Similar Similar Similar Similar Similar Bond length N - Fe 2.02A 2.11A 2.15A 2.32 2.14A Bond length Fe – E7 5.80A 5.71A 5.56A 5.25A 5.25A Bond length Fe – F8 2.15A 2.25A 2.11A 2.21A 2.21A His E7 His F8 Fe N
- 9. Possible ResearchQuestion Data Collection using 3D modelling Data Collection using Database Click here Jmol Click here PyMol Click here NCBI Click here UCSC Click here Ensembl Structuralsimilarity and differences bet Hemoglobin and Myoglobin Myoglobin hemoglobin vs Chimera Swiss PDB Evaluationand Limitationusing 3D modelling Must use a variety of sources/programme to verify/validate the validity and reliability of data collected Average is computed from diff software and checked with database to confirm. Check on methodological limitation using 3D model. (MUST perform 3D Optimization to most stable form structure. Critical and skeptical of result produced by computational chemistry. Major limitation of computation, they assume non-interacting molecule. (Ideal situation, ex molecule in vacuum or isolated state) Most appropriate molecule are those whose coordinates are not theoretical but derive from experimental structural determination (using X ray diffraction) Be careful of predicted arrangement from simulation /3D model Data sources are supported using diff method/3D model/database Certain database like NIST and CRC are more reliable source Check if there is a good agreement bet CRC, diff databases and 3D model prediction before making conclusion Computation programme is always based on approximation and we cannot conclusive prove anything Reflect of validity and reliability of data Is model a true representation of reality? - What is the distance bet Fe and His E7 and F8, and are they the same for diff heme found in hemoglobin/myoglobin - Is His E7/F8 orientation similar for Oxy, Deoxy and Myoglobin. - Is there any differencesbet distance/position/orientationof porphyrin ring for Hemoglobinand Myoglobin - How is Fe2+located, along or out of plane for Hemo/Myoglobin - Is the distance bet Fe and ligand N of porphyrin the same for Hemoglobin/Myoglobin - Is structure/sizeof porphyrin ring same for α and β chain - Is there any variation in terms of Fe and His E7/F8 for fetal hemoglobin and sickle cell hemoglobin - Why His E7 and F8 are locatedin such a way across many different species? Are their orientation highly conserved and why ?
- 10. - Porphyrin gp of heterocyclic made of 4 pyrrole subunit - Porphyrin macrocyclehas 26 (delocalized)pi electron, obey Hückel rule - It is aromatic,4n+2 π. (Highly conjugated system) Heme PorphyrinHeme = Fe + porphyrins ring Heme Heme A Heme B Heme C Mitochondria - cytochromec oxidase - electrontransport O2 Heme = Fe + porphyrin ring – carry O2 Fe2+ located Most abundant Hemoglobin and Myoglobin Mitochondria - cytochromec - electrontransport Fetal Hemoglobin (2α22γ2) Human Hemoglobin (2α2 2β2) Sickle cell Hemoglobin (2α22βS 2) Myoglobin 1 α chain Carbaminohemoglobin Carboxyhemoglobin Oxyhemoglobin
- 11. Heme Heme A Heme B Heme C Mitochondria - cytochromec oxidase - electrontransport Most abundant Hemoglobin and Myoglobin Mitochondria - cytochromec - electrontransport Fetal Hemoglobin (2α22γ2) Human Hemoglobin (2α2 2β2) Sickle cell Hemo (2α22βS 2) Myoglobin 1 α chain Cytochrome Heme in cytochrome,highly conjugated ring sys surrounding Fe Cytochrome- REDOX rxn – mitochondria – ATP/energy productionvia elec transport chain Many type cytochromes – Cyto a, b, c1, a3 Cytochromec, an ancient protein, developed early in the evolution of life. Essential protein for energy/ATP HIGHLY CONSERVED has changed little in millions of years. Many variation – but structureremain relatively unchanged Fe in cytochrome Fe in cytochrome Cytochrome c – heme c
- 12. Hemoglobin A - 2 alpha and 2 beta chains Hemoglobin A2 - 2 alpha and 2 delta chains Hemoglobin F - 2 alpha and 2 gamma chainsHeme (porphyrin) bind to Fe2+using 4 nitrogen atom (histidine gp) Porphyrin - as electron-pair donor - polydentate ligand Fe form 2 additional bonds, one on each side of the heme plane. These binding sites call fifth and sixth coordination sites. This hisitidine is referred as proximal Histidine F8 The sixth coordination site bind oxygen with His E7 nearby Deoxyhemoglobin Fe2+- out plane Can’t fit the ring Heme Hemoglobin - 4 chain - 4 heme (porphyrin) - 4 Fe 2+ Fe2+ Heme (porphyrin) Oxyhemoglobin Fe2+- located in plane Fit the ring Deoxyhemoglobin Fe2+ out plane Can’t fit the ring Out by 0.06nm Fe2+ in plane Fit the ring Human hemoglobin - 2 alpha chain - 144 amino acid - 2 beta chain - 146 amino acid Fe bind to six ligand. 4 with N atom of porphyrin Fifth ligand is donated by His F8 O2 add to Fe as sixth ligand O2 tilt relative to perpendicular of heme plane His F8His F8 His F8 His E7 His E7 His E7 Oxyhemoglobin
- 13. Heme Hemoglobin - 4 chain - 4 heme (porphyrin) - 4 Fe 2+ Fe2+ Heme (porphyrin) Hemoglobin - 2 alpha chain - 144 amino acid - 2 beta chain - 146 amino acid Fe bind to six ligand. 4 with N atom of porphyrin Fifth ligand is donated by His F8 O2 add to Fe as sixth ligand O2 tilt relative to perpendicularof heme plane His E7 locateover Fe, force CO to bind to Fe at an angle. This steric hinderance reduce afinity of CO in hemoglobin. O2 bind to Fe at an angle, its binding not affected by presence of His E7. His (E7) His (F8) vs Myoglobin - 1 chain – 1 heme (porphyrin) - 1 Fe2+ - 154 amino acids HemoglobinMyoglobin Fe2+ Heme (porphyrin) His (F8) His (E7)
