IB Chemistry on HNMR Spectroscopy and Spin spin coupling
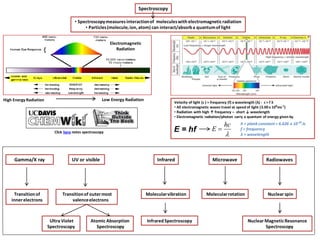
- 1. • Spectroscopymeasuresinteractionof moleculeswith electromagneticradiation • Particles(molecule,ion, atom) can interact/absorba quantumof light Spectroscopy Electromagnetic Radiation Nuclear spin High Energy Radiation Gamma/X ray Transitionof innerelectrons UV or visible Transitionof outermost valenceelectrons Infrared Molecularvibration Microwave Molecularrotation Radiowaves Low Energy Radiation InfraredSpectroscopy Nuclear MagneticResonance Spectroscopy Ultra Violet Spectroscopy Atomic Absorption Spectroscopy Velocity of light (c ) = frequency (f) x wavelength (λ) - c = f λ • All electromagnetic waves travel at speed of light (3.00 x 108 ms-1 ) • Radiation with high ↑ frequency – short ↓ wavelength • Electromagnetic radiation/photon carry a quantum of energy given by E = hf hc E h = plank constant = 6.626 x 10-34 Js f = frequency λ = wavelength Click here notes spectroscopy
- 2. ElectromagneticRadiationand Spectroscopy Radiowaves Nuclear spin Nuclear MagneticResonance Spectroscopy • Organic structure determination • MRI and body scanning Infrared Molecularvibration InfraredSpectroscopy UV or visible Transitionof outervalenceelectron • Organic structure determination • Functional gp determination • Measure bond strength • Measure degree unsaturationin fat • Measure level of alcohol in breath ElectromagneticRadiation UV Spectroscopy Atomic A Spectroscopy • Quantification of metal ions • Detection of metal in various samples ElectromagneticRadiation Interact with Matter (Atoms, Molecules)= Spectroscopy
- 3. Nuclear Magnetic Resonance Spectroscopy (NMR) • Involvenucleus(proton+ neutron)NOT electron • Proton+ neutron= Nucleons • Nucleonslike electronhave spin and magneticmoment (acts like tiny magnet) Nuclei with even number of nucleon (12C and 16O) • Even numberof protonand neutron– NO net spin • Nucleonspin cancel out each other –Nucleushave NO overallmagneticmoment – NOT absorb radiowave Nuclei with odd number of nucleon (1H, 13C, 19F, 31P) -Nucleonhave net spin – Nucleushave NET magneticmoment – Absorbradiowave • Nuclei with net spin – magnetic momentwill interactwith radiowaves • Nuclei have a “spin”associated with them (i.e., they act as if they were spinningaboutan axis) due to the spin associatedwith theirprotonand neutron. • Nuclei are positivelycharged,theirspin inducesa magnetic field • NMR spectroscopy does not work for nuclei with even numberof protons and neutrons - nuclei have no net spin. Nuclear Magnetic Resonance Spectroscopy (NMR) Spin cancel each other Net spin
- 4. Main features of HNMR Spectra 1. Number of diff absorption peak – Number of diff proton/chemicalenvironment 2. Area under peak - Number of hydrogen in a particularproton/chemicalenvironment(Integration trace) - Ratio of number of hydrogen in each environment 3. Chemicalshift - Chemicalenvironment where proton is in - Spinning electron create own magnetic field, creating a shielding effect - Proton which are shielded appear upfield. (Lower frequency for resonance to occur) - Proton which are deshielded appear downfield. (Higher frequency for resonance to occur) - Measured in ppm (δ) 4. Splitting pattern - Due to spin-spin coupling - Number of peak split is equal to number of hydrogen on neighbouring carbon+1 (n+1) peak Chemical ShiftNMR spectrum CH3CH2Br Number of peaks Area under peaks Chemical shift Splitting pattern Nuclear Magnetic Resonance Spectroscopy (NMR) Click here khan NMR videos.
- 5. NMR spectrum CH3CH2Br Number of peaks Splitting pattern Click here khan NMR videos. Number equi H Multiplicity Ratio 0 Singlet 1 1 Doublet 1: 1 2 Triplet 1 : 2 : 1 3 Quartet 1 : 3 : 3 : 1 4 Quintet 1 : 4 : 6 : 4 : 1 5 Hextet 1 : 5 : 10 : 10 : 5 : 1 6 Septet 1 : 6 : 15 :20 : 15 : 6 : 1 SplittingPattern Singlet – Neighbouring Carbon with No H Doublet – Neighbouring Carbon with 1H Triplet – Neighbouring Carbon with 2H Quartet – Neighbouring Carbon with 3H • Equiv H in same chemicalenvironment have no splitting effect on each other • Equiv H do not split each other • All Equiv H in same chemicalenvironment will produce a same peak • Spin spin coupling – occurwhen proton have diff chemicalshift • Splitting not observed for proton that are chemicallyequivalent/samechemicalshift High Resolution (NMR) Main features of HNMR Spectra 1. Number of diff absorption peak – Number of diff proton/chemicalenvironment 2. Area under peak - Number of hydrogen in a particularproton/chemicalenvironment(Integration trace) - Ratio of number of hydrogen in each environment 3. Chemicalshift - Chemicalenvironment where proton is in - Spinning electron create own magnetic field, creating a shielding effect - Proton which are shielded appear upfield. (Lower frequency for resonance to occur) - Proton which are deshielded appear downfield. (Higher frequency for resonance to occur) - Measured in ppm (δ) 4. Splitting pattern - Due to spin-spin coupling - Number of peak split is equal to number of hydrogen on neighbouring carbon+1 (n+1) peak
- 6. Br ׀ H – C – Br ׀ H – C – H ׀ H (n + 1 rule) • Equiv H in same environment do not split each other. • If H has n equiv proton on neighboring carbon, signal for H will split to n + 1 peak. • H nuclei split neighbouring H in CH3 to 2 peak, (doublet). H nuclei split CH3 methyl gp to doublet • H can align with EMF or against EMF. • CH3 will experience2 diff EMF • One lower, one higher EMF • Split to doublet EMF align against MF produce by H • Overall MF experience CH3 lower • H from CH3 will absorb at lower freq (upfield) EMF EMF align with MF produce by H • Overall MF experience by CH3 higher • H from CH3 will absorb at higher freq (downfield) EMF • CH3 spilt to doublet by 1 adj H • CH3 experience2 slightly diff MF due to neighbouring H MF MF Split with relative intensity of 1 : 1 Downfield Upfield High Resolution (NMR) Br ׀ H – C – Br ׀ H – C – H ׀ H Br ׀ H – C – Br ׀ H – C – H ׀ H Br ׀ H – C – Br ׀ H – C – H ׀ H doublet Splitting peaks occuras effective MF experience by H nuclei is modified by MF produced by neighbouring proton/H
- 7. (n + 1 rule) • If H has n equiv proton on neighboring carbon, signal for H, split to n + 1 peak. • 2H nuclei split neighbouring H in CH3 to 3 peak, (triplet). 2H nuclei split CH3 methyl to triplet • H can align with EMF or against EMF. • CH3 will experience 3 diff EMF • One lower, one higher , one no net change • Split to triplet (ratio 1 : 2 : 1 ) EMF align against MF by H • Both align against EMF (Net lower EMF) •Overall MF experience by CH3 lower • H from CH3, absorb lower freq EMF EMF align with MF produce by H • Both H align with EMF (Net greater EMF) • Overall MF experience by CH3 higher • H from CH3, absorb at higher freq EMF EMF MF MF MF MF EMF align with/against MF produce by H • 1 align with and 1 against EMF • MF by H cancel each other • Overall MF experience by CH3 the same Split with relative intensity of 1 : 2 : 1 • CH3 spilt to triplet by 2 adj H • CH3 experience3 diff MF due to 2 adjacentH Downfield Upfield H – C – H ׀ H – C – H ׀ H H – C – H ׀ H – C – H ׀ H H – C – H ׀ H – C – H ׀ H H – C – H ׀ H – C – H ׀ H H – C – H ׀ H – C – H ׀ H Br ׀ H – C – H ׀ H – C – H ׀ H triplet Splitting peaks occur as effective MF experience by H nuclei is modified by MF produced by neighbouring proton/H High Resolution (NMR)
- 8. 3H nuclei split CH2 methylene to quartet • H can align with EMF or against EMF. • CH2 will experience 4 diff EMF • Split to quartet (ratio 1 : 3 : 3 : 1 ) EMF align against MF by H • 3 H align against EMF (Lower EMF) •Overall MF experience CH2 lower • H from CH2, absorb at lower freq EMF EMF align with MF by H • 3 H align with EMF (Net greater EMF) • Overall MF experience by CH2 higher • H from CH2, absorb at higher freq EMF EMF MF MF EMF align with/against MF by H • 2 align with and 1 against EMF (higher) • 2 align against and 1 with EMF (lower) • 2 diff MF experience by CH2 in 3 : 3 ratio Split with relative intensity of 1 : 3 : 3 : 1 • CH2 spilt to quartet by 3 adjacent H • CH2 experience4 diff MF due to 3 adjacent H (n + 1 rule) • If H has n equiv protons on neighboring carbons, signal for H, split to n + 1 peak. • 3H nuclei split neighbouring H in CH3 into 4 peak, called quartet. H – C – H ׀ H – C – H ׀ H H – C – H ׀ H – C – H ׀ H H – C – H ׀ H – C – H ׀ H H – C – H ׀ H – C – H ׀ H H – C – H ׀ H – C – H ׀ H H – C – H ׀ H – C – H ׀ H H – C – H ׀ H – C – H ׀ H H – C – H ׀ H – C – H ׀ H H – C – H ׀ H – C – H ׀ H H – C – H ׀ H – C – H ׀ H quartet Splitting peaks occur as effective MF experience by H nuclei is modified by MF produced by neighbouring proton/H High Resolution (NMR)
- 9. High Resolution (NMR) Singlet peak • H nuclei attachto electronegativeatom, O - NO splitting – Singlet • H nuclei attachto neighbouring C without any H - NO splitting – Singlet • Equiv H nuclei do not split each other but will split neighbouring H • CH3 spilt to triplet by 2 adj H • CH3 experience3 diff MF due to 2 adj H • CH2 spilt to quartet by 3 adj H • CH2 experience4 diff MF due to 3 adj H • No signal splitting from coupling bet hydroxyl proton and methylene proton of CH2 – despite 2 adjacent H • H attached to O, undergo rapid chemical exchange, transfer rapidly from each other /loss of H • Spin coupling due to H (OH) on methylene proton CH2 is negligible/not seen. • NO triplet split on OH due to 2 adjacent H from CH2 – Only singlet H H ׀ ׀ HO- C- C- H ׀ ׀ H H CH3 • chemicalshift ≈ 1 • integration = 3 H • split into 3 CH2 • chemicalshift ≈ 3.8 • integration = 2 H • split to 4 OH • chemicalshift ≈ 4.8 • integration = 1 H • No split (Singlet) 3 21 Triplet splitQuartet split Singlet split tripletquartet
- 10. - Equiv H in same chemicalenvironment have no splitting effect on each other - All Equiv H produce same signal O ‖ CH3-C-O-CH2-CH3 HO-CH2-CH3 O ‖ HO-C-CH2-CH3 O ‖ CH3-C-CH2-CH2-CH3 Equivalent Hydrogen in same chemical Environment (chemical Shift) 4 diff chemicalenvironment • 4 peak ratio 3:2:3:2 3 diff chemicalenvironment • 3 peak ratio 3:2:1 3 diff chemicalenvironment • 3 peak ratio 3:3:2 3 diff chemicalenvironment • 3 peak ratio 3:2:1 12 33232 32132 3 2 equiv H 3 equiv H 3 equiv H 2 equiv H 21 3 equiv H2 equiv H1 equiv H 3 equiv H2 equiv H 3 equiv H 3 equiv H 2 equiv H 1 equiv H
- 11. Equivalent Hydrogen in molecule with plane of symmetry Equiv H - Hydrogen attach to carbon in particularchemicalenvironment • Equiv H in same environment have no splitting effect on each other • H on neighbouring carboncan be equiv if they are in same environment • All Equiv H in same environment will produce a same signal. CH3 | CH3 – C -CH3 | CH3 1 chemicalenvironment • 1 peak O ║ CH3-CH2-C-CH2- CH3 2 diff chemicalenvironment • 2 peak ratio 3:2 CI ׀ CH3-C-CH3 ׀ H CH3 ׀ HO-CH2- C- H ׀ CH3 2 4 2 3212 612 161 12 equiv H 2 diff chemicalenvironment • 2 peak ratio 6:1 4 diff chemicalenvironment • 4 peak ratio 6:1:1:2 1 equiv H 6 equiv H 6 equiv H1 equiv H2 equiv H 3 equiv H2 equiv H
- 12. O CH3 ‖ ׀ H-C – C-CH3 ׀ CH3 CH3 | H-C-OH | CH3 O CH3 ‖ ׀ CH3-C-O-C-H ׀ CH3 H CH3 ׀ ׀ CI- C – C- CH3 ׀ ׀ H CH3 9.7 91611 92631 Equivalent Hydrogen in molecule with plane of symmetry Equiv H - Hydrogen attach to carbon in particularchemicalenvironment • Equiv H in same environment have no splitting effect on each other • H on neighbouring carboncan be equiv if they are in same environment • All Equiv H in same environment will produce a same signal. 3 diff chemicalenvironment • 3 peak ratio 6:1:1 2 diff chemicalenvironment • 2 peak ratio 9:1 1 equiv H 6 equiv H 1 equiv H 9 equiv H1 equiv H 3 diff chemicalenvironment • 3 peak ratio 6:3:1 2 diff chemicalenvironment • 2 peak ratio 9:2 6 equiv H 3 equiv H 1 equiv H 9 equiv H 2 equiv H
- 13. CI CI | | C = C | | H H CI CI CI ׀ ׀ ׀ H- C- C - C- H ׀ ׀ ׀ CI H CI H H ׀ ׀ CI- C – C- CI ׀ ׀ H H H H ׀ ׀ H – C – C - H ׀ ׀ H H 4.56.1 22 1 64 Equivalent Hydrogen in molecule with plane of symmetry Equiv H - Hydrogen attach to carbon in particularchemicalenvironment • Equiv H in same environment have no splitting effect on each other • H on neighbouring carboncan be equiv if they are in same environment • All Equiv H in same environment will produce a same signal. 2 diff chemicalenvironment • 2 peak ratio 1:2 1 chemicalenvironment • 1 peak 2 equiv H 1 equiv H 2 equiv H 1 chemicalenvironment • 1 peak 4 equiv H 1 chemicalenvironment • 1 peak 6 equiv H
- 14. O ‖ CH3-C-O-CH2CH3 HO-CH2CH3 O ‖ HO-C-CH2CH3 O ‖ CH3-C-CH2CH2CH3 12 • Equiv H in same environment have no splitting effect on each other • Equiv H do not split each other • All equiv H in same environment will produce a same peak . Triplet 2 adj H Septet 5 adj H Singlet No adj H Triplet 2 adj H Triplet 2 adj H Quartet 3 adj H Singlet OH – No split Triplet 2 adj H Singlet No adj H Quartet 3 adj H Triplet 2 adj H Quartet 3 adj HSinglet No adj H Splitting Pattern by neighbouring H 3213232 321332 4 chemicalenvironment • 4 peak ratio 3:2:3:2 3 chemicalenvironment • 3 peak ratio 3:2:1 3 chemicalenvironment • 3 peak ratio 3:3:2 3 chemicalenvironment • 3 peak ratio 3:2:1
- 15. CH3 ׀ HO-CH2–C-H ׀ CH3 CH3 ׀ CH3 – C – CH3 ׀ CH3 O ‖ CH3CH2-C-CH2CH3 CI ׀ CH3- C – CH3 ׀ H 2 4 Singlet No adj H Triplet 2 adj H Quartet 3 adj H Doublet 1 adj H Heptet 6 adj H Doublet 1 adj H Doublet 1 adj H Singlet OH- No split Nonet 8 adj H 3212 611261 Splitting Pattern by neighbouring H • Equiv H in same environment have no splitting effect on each other • Equiv H do not split each other • All equiv H in same environment will produce a same peak . 2 chemicalenvironment • 2 peak ratio 6:1 1 chemical environment • 1 peak 2 chemicalenvironment • 2 peak ratio 3:2 4 chemicalenvironment • 4 peak ratio 6:1:1:2
- 16. O CH3 ‖ ׀ H-C- C-CH3 ׀ CH3 CH3 ׀ H-C –OH ׀ CH3 O CH3 ‖ ׀ CH3-C-O-C – H ׀ CH3 H CH3 ׀ ׀ CI- C – C –CH3 ׀ ׀ H CH3 9.7 Heptet 6 adj H Singlet OH- No split Doublet 1 adj H Singlet No adj H Doublet 1 adj H Heptet 6 adj H Singlet No adj H Singlet No adj H Singlet No adj H Singlet No adj H 91611 9261 3 Splitting Pattern by neighbouring H • Equiv H in same environment have no splitting effect on each other • Equiv H do not split each other • All equiv H in same environment will produce a same peak . 3 chemicalenvironment • 3 peak ratio 6:1:1 2 chemical environment • 2 peak ratio 9:1 3 chemicalenvironment • 3 peak ratio 6:3:1 2 chemical environment • 2 peak ratio 9:2
- 17. Singlet Splitting Pattern • Equiv H in same environment have no splitting effect on each other • All equiv H in the same environment will produce a same peak . • Singlet can be due to presence of OH or no adjacentH CH3 ׀ CH3 – C – CH3 ׀ CH3 Singlet No adj H O CH3 ‖ ׀ H-C- C – CH3 ׀ CH3 Singlet No adj H 9.7 Singlet No adj H H CH3 ׀ ׀ CI- C – C- CH3 ׀ ׀ H CH3 Singlet No adj H Singlet No adj H H H ׀ ׀ CI- C – C – CI ׀׀ H H Singlet No adj H 924 12 91 Singlet due to • Equiv H in same environ • No adj H Singlet due to • Equiv H in same environ • No adj H Singlet due to • Equiv H in same environ • Equiv H do not split each other Singlet due to • Equiv H in same environ • No adj H
- 18. Singlet All equiv H Singlet No adj H Singlet No adj H H H ׀ ׀ H – C –C- H ׀ ׀ H H CH3 ׀ CH3O – C – CH3 ׀ CH3 O ‖ HO-C-CH3 12 Singlet No adj H 2 Singlet No adj H O ‖ HO-C-H Singlet No adj H 10.6 8.3 Singlet No adj H 3111 6 93 Singlet Splitting Pattern • Equiv H in same environment have no splitting effect on each other • All equiv H in same environment will produce a same peak . • Singlet can be due to presence of OH or no adjacentH Singlet due to • Equiv H in same environ • Equiv H do not split each other Singlet due to • Equiv H in same environ • No adj H Singlet due to • OH in COOH • H in CHO Singlet due to • OH in COOH • No adj H
- 19. 2 diff proton environment, Ratio H – 3: 5 • Peak A – No split (No H on adj C) • Peak B – split to 3 (2H on adj C) • Peak C – split to 3 (2H on adj C) • Peak D – split to 2 (1H on adj C) A B 3 5 2 1 2 C D 7.38 All H in benzene consider • as 1 proton environment 7.38 2 E 1 D 2 5 C 2 32 A B 3 diff proton environment, Ratio H – 3: 2 : 5 • Peak A – split to 3 (2H on adj C) • Peak B – split to 4 (3H on adj C) • Peak C – split to 3 (2H on adj C) • Peak D – split to 3 (2H on adj C) • Peak E – split to 2 (1H on adj C) Moleculewith benzenering Moleculewith benzenering All H in benzene consider • as 1 proton environment High Resolution (NMR)
- 20. A C 3 5 2 1 2 D E 7.38 7.38 2 F 1 E 2 5 D 3 12 AB 4 diff environment, Ratio H – 1 : 2 : 2 : 5 • Peak A – No split for OH • Peak B – split to 3 (2H on adj C) • Peak C – split to 3 (2H on adj C) • Peak D – split to 3 (2H on adj C) • Peak E – split to 3 (2H on adj C) • Peak F – split to 2 (1H on adj C) 2 B 3 diff proton environment, Ratio H – 3 : 2 : 5 • Peak A – split to 3 (2H on adj C) • Peak B – split to 4 (3H on adj C) • Peak C – split to 3 (2H on adj C) • Peak D – split to 3 (2H on adj C) • Peak E – split to 2 (1H on adj C) 3 4 C 2 High Resolution (NMR) Moleculewith benzenering All H in benzene consider • as 1 proton environment All H in benzene consider • as 1 proton environment Moleculewith benzenering
- 21. A C 6 5 2 1 2 D E 7.38 1 B 3 diff environment, Ratio H – 6 : 1 : 5 • Peak A – split to 2 (1H on adj C) • Peak B – split to 7 (6H on adj C) • Peak C – split to 3 (2H on adj C) • Peak D – split to 3 (2H on adj C) • Peak E – split to 2 (1H on adj C) 5 High Resolution (NMR) Moleculewith benzenering All H in benzene consider • as 1 proton environment Unknown X have MF of C3H6O2 with HNMR shown Chemical shift/ppm Number H atoms Splitting pattern 1.3 3 3 4.3 2 4 8 1 1 i. Deduce IHD 32 8 4.3 1.3 Triplet 2 adj H Quartet 3 adj H Singlet No adj H 1 3 diff environment • 3 peak/chemical shift O ‖ HO-C-CH2-CH3 yx HC 1 2 )12262( 2 22 IHD IHD yx IHD ii. Deduce molecular structure Molecule Index H2 Deficiency C3H6O2 1 IHD = 1 (1 double bond or ring) H in COOH – singlet – 8 ppm (next to COO) - No adj H bond neighbour C H in CH3 - triplet – 1.3 ppm (next to CH2) - 2 adj H bond neighbour C H in CH2 - quartet – 4.3 ppm (next to C=O) - 3 adj H bond neighbour C O ‖ HO-C-CH2CH3
- 22. Unknown X have mass composition of 85.6%C, 14.4%H Mass spectra, IR and NMR shown below. % composition mass C 85.6 H 14.4 m/e IB Question 10 20 30 40 50 60 70 80 90 i. Deduce EF and MF of compound abundance 28 42 56 84 2 1 0 Singlet All equiv H 12 Empirical formula = CH2 Molecular ion, M+ = RMM = 84 n(EF) = MF n(CH2)= 84 n (12 + 2.01) = 84 n = 6 MF = C6H12 C – H stretch (2840 – 3000) C – H bend (1200) Mass spec IR spec HNMR spec yx HC 1 2 )12262( 2 22 IHD IHD yx IHD Molecule Index H2 Deficiency C6H12 1 M+ peak M+ = C6H12 + = 84 ii. Deduce IHD of compound IHD = 1 (1 double bond or ring) iii. Deduce the molecular structure IR spec → C-H stretch and C – H bend No C=C absorption at 1610 No C =O/C-O/OH functional gp HNMR spec → 1 singlet peak All equiv H at same chemical environment MolecularFormula H in CH2- singlet – 1 ppm All 12 H in same chemical environment (symmetry)
- 23. O ‖ CH3C-OH % composition mass C 40 H 6.7 O 53.3 m/e IB Question i. Deduce EF and MF of compound abundance 28 45 60 2 Singlet No adj H 3 Empirical formula = CH2O Molecular ion, M+ = RMM = 60 n(EF) = MF n(CH2O)= 60 n (12 + 2.01 + 16) = 60 n = 2 MF = C2H4O2 C – H stretch (2840 – 3000) C – H bend (1200) yx HC 1 2 )4222( 2 22 IHD IHD yx IHD Molecule Index H2 Deficiency C2H4O2 1 M+ peak M+ = C2H4O2 + = 60 ii. Deduce IHD of compound IHD = 1 (1 double bond or ring) iii. Deduce the molecular structure IR spec → C-H /O-H stretch (broad absorption) C=O absorption at 1680 C-O absorption at 1200 C=O/C-O/OH functional gp HNMR spec → 2 singlet peak 2 peak/diff environment, ratio 3 : 1 MolecularFormula Unknown X mass composition of 40% C, 6.7%H, 53.3%O Mass spectra, IR and NMR shown below. 10 20 30 40 50 60 70 80 90 15 17 O – H stretch (3230 -3550) C – O stretch (1000-1300)C = O stretch (1680 -1740) Singlet No adj H 12 1 C – H stretch (2840 – 3000) O ‖ CH3C-OH H in COOH – singlet – 12ppm (next to COO) H in CH3 - singlet – 2 ppm (next to C=O)
- 24. m/e IB Question i. Deduce structural formula X 29 45 74 1 3 Molecular ion, M+ = RMM = 74 MF = C3H6O2 C – H stretch (2840 – 3000) C – H bend (1200) yx HC 1 2 )6232( 2 22 IHD IHD yx IHD Molecule Index H2 Deficiency C3H6O2 1 M+ peak M+ = C3H6O2 + = 74 ii. Deduce IHD of compound IHD = 1 (1 double bond or ring) iii. Deduce the molecular structure IR spec → C-H /O-H stretch (broad absorption) C=O absorption at 1680 C-O absorption at 1200 C=O/C-O/OH functional gp HNMR spec → triplet/quartet – CH3CH2 present → singlet – at 11ppm - COOH 3 peak/diff environment, ratio 3:2:1 MolecularFormula 10 20 30 40 50 60 70 80 90 15 17 O – H stretch (3230 -3550) C – O stretch (1000-1300)C = O stretch (1680 -1740) Singlet No adj H 11 1 C – H stretch (2840 – 3000) Unknown X have MF C3H6O2 Mass spectra, IR and NMR shown below. O ‖ CH3CH2-COH 2 2 Triplet 2 adj H Quartet 3 adj H C2H5 + = 29 COOH+ = 45 CH3 + = 15 OH+ = 17 O ‖ HO-C-CH2-CH3 H in COOH – singlet – 11ppm (next to COO) H in CH2 - quartet – 2 ppm (next to CH3) H in CH3 - triplet – 1 ppm (next to CH2) O ‖ CH3CH2-C-OH CH3 + = 15 C2H5 + = 29 COOH+ = 45
- 25. H O H H ׀ ‖ ׀ ׀ H - C – C –O – C – C– H ׀ ׀ ׀ H H H H O H H ׀ ‖ ׀ ׀ H - C – C –O – C – C– H ׀ ׀ ׀ H H H % composition mass C 40 H 6.7 O 53.3 m/e IB Question i. Deduce EF and MF of compound 29 45 88 1 3 Empirical formula = C2H4O Molecular ion peak, M+ = RMM = 88 n(EF) = MF n(C2H4O)=88 n (24 + 1.01 x 4 + 16) = 88 n = 2 MF = C4H8O2 C – H stretch (2840 – 3000) yx HC 1 2 )8242( 2 22 IHD IHD yx IHD Molecule Index H2 Deficiency C4H8O2 1 M+ peak M+ = C4H8O2 + = 88 ii. Deduce IHD of compound IHD = 1 (1 double bond or ring) iii. Deduce the molecular structure IR spec → No O-H stretch (No broad absorption) C=O absorption at 1680 C-O absorption at 1200 C=O/C-O, functional gp HNMR spec → triplet/quartet – CH3CH2 present → singlet – at 2 ppm – CH3 next to C=O 3 peak/diff environment, ratio 3:3:2 MolecularFormula 10 20 30 40 50 60 70 80 90 15 C – O stretch (1000-1300)C = O stretch (1680 -1740) 4 2 3 2 Triplet 2 adj H Quartet 3 adj H C2H5 + = 29 C2H5O+ = 45 CH3 + = 15 CH3CO+ = 43 H in CH3 - triplet – 1 ppm (next to CH2) H in CH3 – singlet – 2 ppm (next to C=O) H in CH2 - quartet – 4 ppm (next to O) Unknown X have EF C2H4O Mass spectra, IR and NMR shown below. 43 Singlet No adj H Ester group
- 26. O H H ‖ ׀ ׀ H–C –O – C – C– H ׀ ׀ H H % composition mass C 48.63 H 8.18 O 43.19 m/e IB Question i. Deduce EF and MF of compound 29 45 74 1 3 Empiricalformula = C3H6O2 Molecular ion peak, M+ = RMM = 74 n(EF) = MF n(C3H6O2)=74 n (12 x 3 + 1.01 x 6 + 16 x 2) = 74 n = 1 MF = C3H6O2 C – H stretch (2840 – 3000) yx HC 1 2 )6232( 2 22 IHD IHD yx IHD Molecule Index H2 Deficiency C3H6O2 1 M+ peak M+ = C3H6O2 + = 74 ii. Deduce IHD of compound IHD = 1 (1 double bond or ring) iii. Deduce the molecular structure IR spec → No O-H stretch (No broad absorption) C=O absorption at 1680 C-O absorption at 1200 C=O/C-O, functional gp HNMR spec → triplet/quartet – CH3CH2 present → singlet – at 8 ppm – H next to COO 3 peak/diff environment, ratio 3: 2: 1 MolecularFormula 10 20 30 40 50 60 70 80 90 15 C – O stretch (1000-1300)C = O stretch (1680 -1740) 8 21 4 Triplet 2 adj H Quartet 3 adj H C2H5 + = 29 C2H5O+ = 45 CH3 + = 15 HCOO+ = 45 H in CHO– singlet – 8 ppm (next to COO) H in CH2 - quartet – 4 ppm (next to O) H in CH3 - triplet – 1 ppm (next to CH2) Singlet No adj H Ester group Unknown X mass composition of 48.63%C, 8.18% H, 43.19%O Mass spectra, IR and NMR shown below. O H H ‖ ׀ ׀ H–C –O – C – C– H ׀ ׀ H H
- 27. % composition mass C 15.4 H 3.24 I 81.36 m/e IB Question i. Deduce EF and MF of compound abundance 29 127 156 3 1 3 Empiricalformula = C2H5I Molecular ion peak, M+ = RMM = 156 n(EF) = MF n(C2H5I)=156 n (12 x 2 + 1.01 x 5 + 127) = 156 n = 1 MF = C2H5I C – H stretch (2840 – 3000) C – H bend (1200) syx XHCMolecule Index H2 Deficiency C2H5I 0 M+ peak M+ = C2H5I+ = 156 ii. Deduce IHD of compound IHD = 0 (Saturated) iii. Deduce the molecular structure IR spec → C-H stretch and C – H bend No C=C absorption at 1610 No C =O/C-O/OH functional gp HNMR spec → triplet/quartet - CH3CH2 present 2 peak/ diff environment, ratio 3:2 MolecularFormula 20 30 40 120 150 Unknown X have mass composition 15.4%C, 3.24%H, 81.36%I Mass spectra, IR and NMR shown below. 0 2 )15222( 2 22 IHD IHD syx IHD C – CI stretch (700-800) H H ׀ ׀ H - C- C - I ׀ ׀ H H C2H5 + = 29 I+ = 127 Triplet 2 adj H Quartet 3 adj H 2 H in CH3 - triplet – 1 ppm (next to CH2) H in CH2 - quartet – 3 ppm (next to I) H H ׀ ׀ I - C- C - H ׀ ׀ H H
- 28. Tetramethyl Silane (TMS) as STD •Strong peak upfield (shielded) •Silicon has lower EN value < carbon • Electron shift to carbon • H in CH3 more shielded • Experience lower EMF, absorb ↓ freq • UPFIELD ≈ 0 Click here for more complicated proton chemical shift • 3 diff proton environment • Ratio of 3:2:1 CH3 • chemicalshift ≈ 1 • integration = 3 H • split to 3 CH2 • chemicalshift ≈ 3.8 • integration = 2 H • split to 4 OH • chemicalshift ≈ 4.8 • integration = 1 H • No split (Singlet) 321 Upfield 12 Advantages using TMS • Volatile and can be removed from sample • All 12 hydrogen in same proton environment • Single strong peak, upfield, wont interfere with other peak • All chemical shift, in ppm (δ) are relative to this STD, ( zero) Nuclear Magnetic Resonance Spectroscopy (HNMR) HO-CH2-CH3 CH3 ׀ H3C – Si – CH3 ׀ CH3 Click here Spectra database (Ohio State) Click here Spectra database (NIST) TMS Downfield
- 29. 1H NMR Spectrum O ‖ HO-C-CH2CH3 3 diff environment, Ratio H - 3:2:3 • Peak A – split to 3 (2H on neighbour C) • Peak B - No split • Peak C – split to 4 (3H on neighbour C) 3 diff environment, Ratio H - 3:2:1 • Peak A – split to 3 (2H on neighbour C) • Peak B – split to 4 (3H on neighbourC) • Peak C – No split A B C B A C 12 32 3 321 O ‖ CH3-C-O-CH2CH3
- 30. O ‖ CH3-C-CH2-CH2-CH3 3 diff environment,Ratio H - 3:2:1 • Peak A – split to 3 (2H on neighbourC) • Peak B – split to 4 (3H on neighbourC) • Peak C – No split 4 diff environment,Ratio H - 3:2:2:3 • Peak A – split to 3 (2H on neighbourC) • Peak B – split to 6 (5H on neighbourC) • Peak C – No split • Peak D – split to 3 (2H on neighbourC) A B C 3 B A C D 21 322 3 HO-CH2-CH3 1H NMR Spectrum
- 31. O ‖ H-C-CH3 4 diff environment, Ratio H – 3:2:2:3 • Peak A – split to 3 (2H on neighbour C) • Peak B – split to 6 (5H on neighbour C) • Peak C – No split • Peak D – split to 3 (2H on neighbour C) ABCD 2 diff environment,Ratio H - 3:1 • Peak A – split to 2 (1H on neighbourC) • Peak B – split to 4 (3H on neighbourC) 9.8 A B 322 3 3 1 O ‖ CH3-C-O-CH2-CH2CH3 1H NMR Spectrum
- 32. 3 diff environment, Ratio H - 6:1:1 • Peak A – split to 2 (1H on neighbour C) • Peak B – No split • Peak C – split to 7 (6H on neighbour C) O CH3 ‖ ׀ CH3-C-O-C-H ׀ CH3 A B C A B C 3 diff environment, Ratio H - 6:3:1 • Peak A – split to 2 (1H on neighbour C) • Peak B – No split • Peak C – split to 7 (6H on neighbour C) Molecule with plane of symmetry 611 631 CH3 ׀ H- C – OH ׀ CH3 Molecule with plane of symmetry 1H NMR Spectrum
- 33. 2 diff environment, Ratio H – 6:4 • Peak A – split to 3 (2H on neighbour C) • Peak B – split to 4 (3H on neighbourC) A B A B 64 91 2 diff environment,Ratio H – 9:1 • Peak A – No split • Peak B – No split Molecule with plane of symmetry O ‖ CH3CH2-C-CH2CH3 Molecule with plane of symmetry O CH3 ‖ ׀ H-C – C – CH3 ׀ CH3 1H NMR Spectrum
- 34. 4 diff environment, Ratio H- 6:1:1:2 • Peak A – split to 2 (1H on neighbour C) • Peak B – split to 7 (6H on neighbourC) • Peak C – No split • Peak D – split to 2 (1H on neighbour C) A B D C 2 diff environment,Ratio H – 6:1 • Peak A – split to 2 (1H on neighbourC) • Peak B – split to 7 (6H on neighbourC) A B 6112 61 Molecule with plane of symmetry CH3 ׀ HO-CH2-C-H ׀ CH3 Molecule with plane of symmetry CH3-CH-CH3 ׀ CI 1H NMR Spectrum
