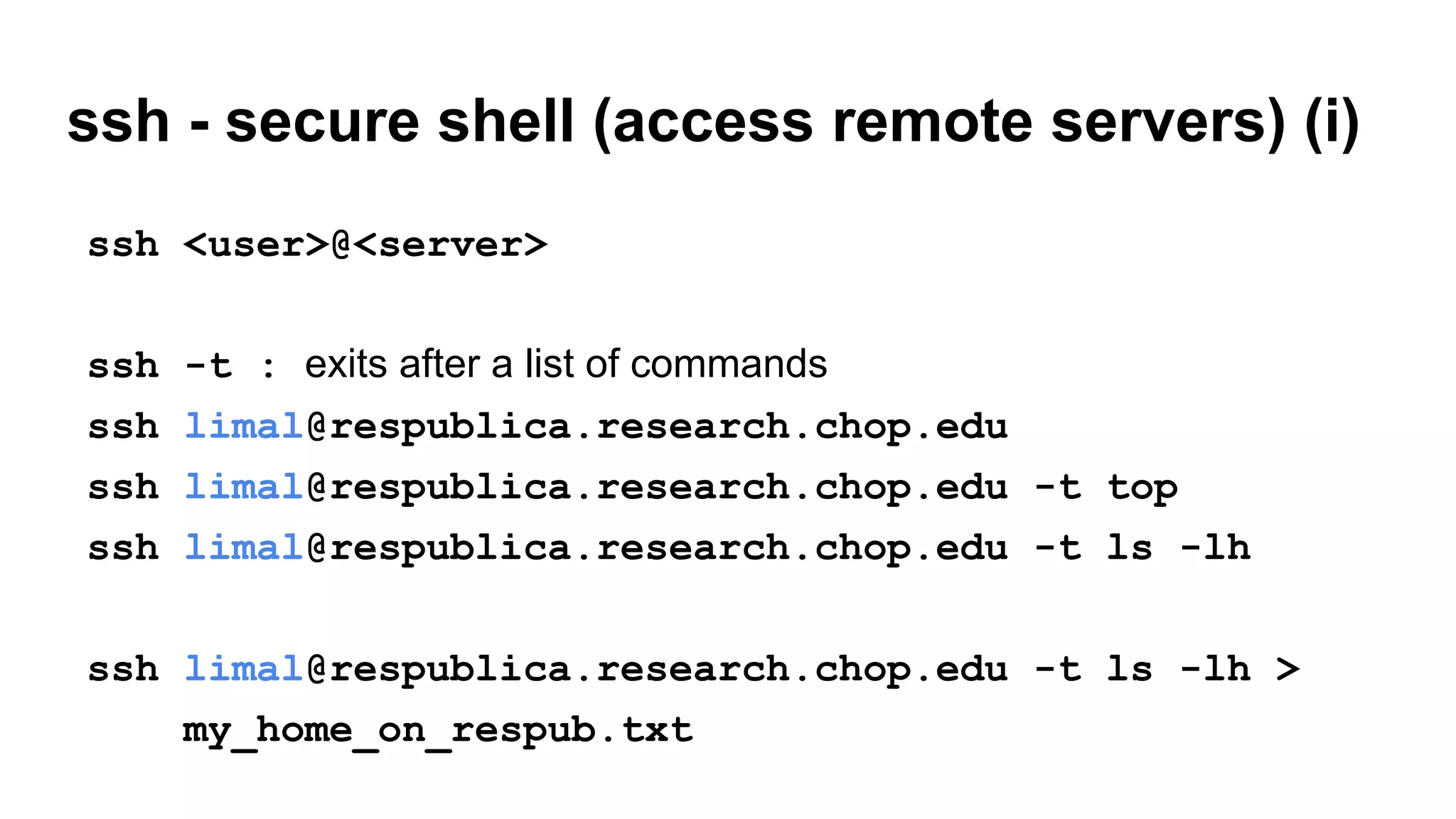
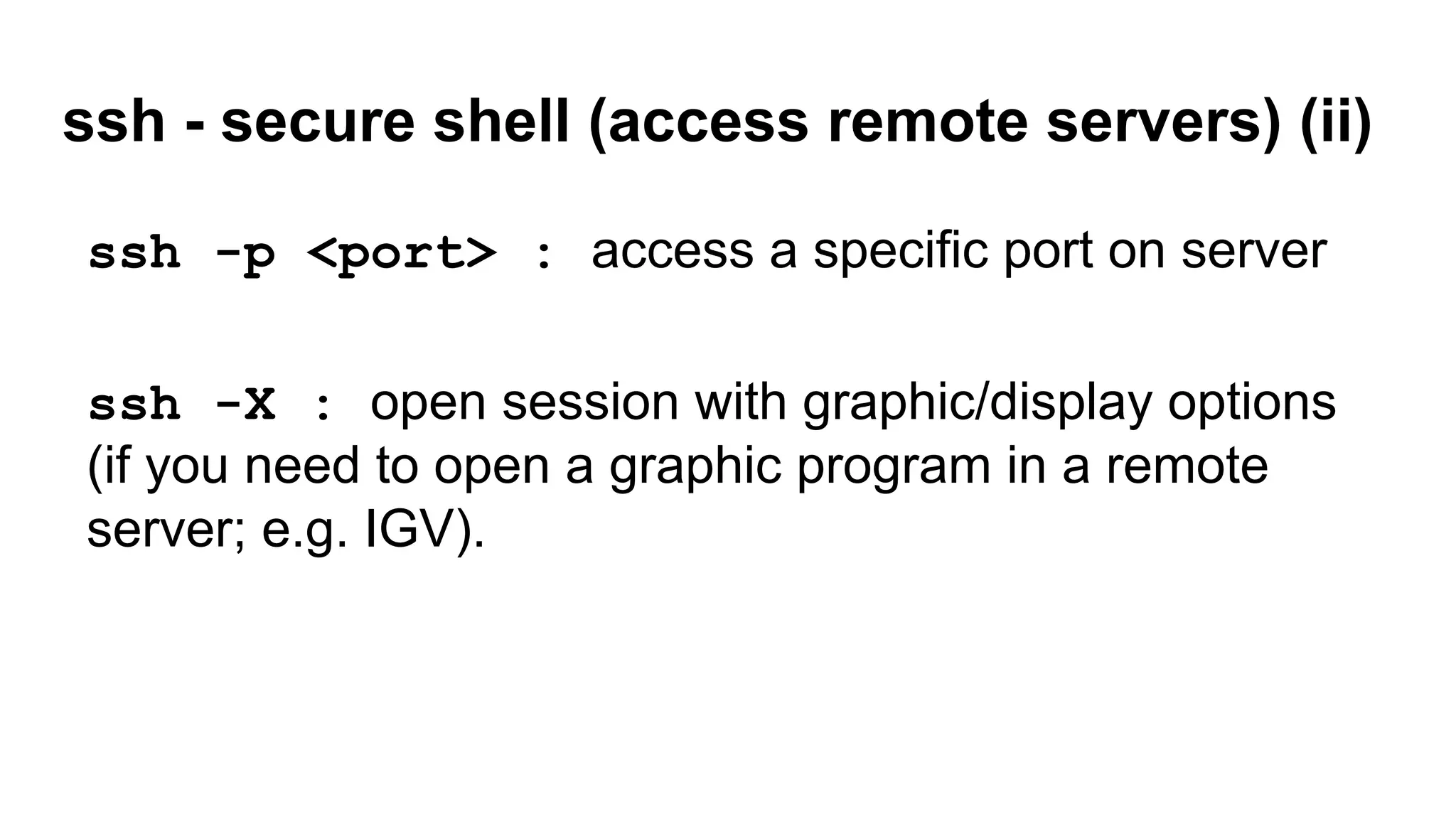
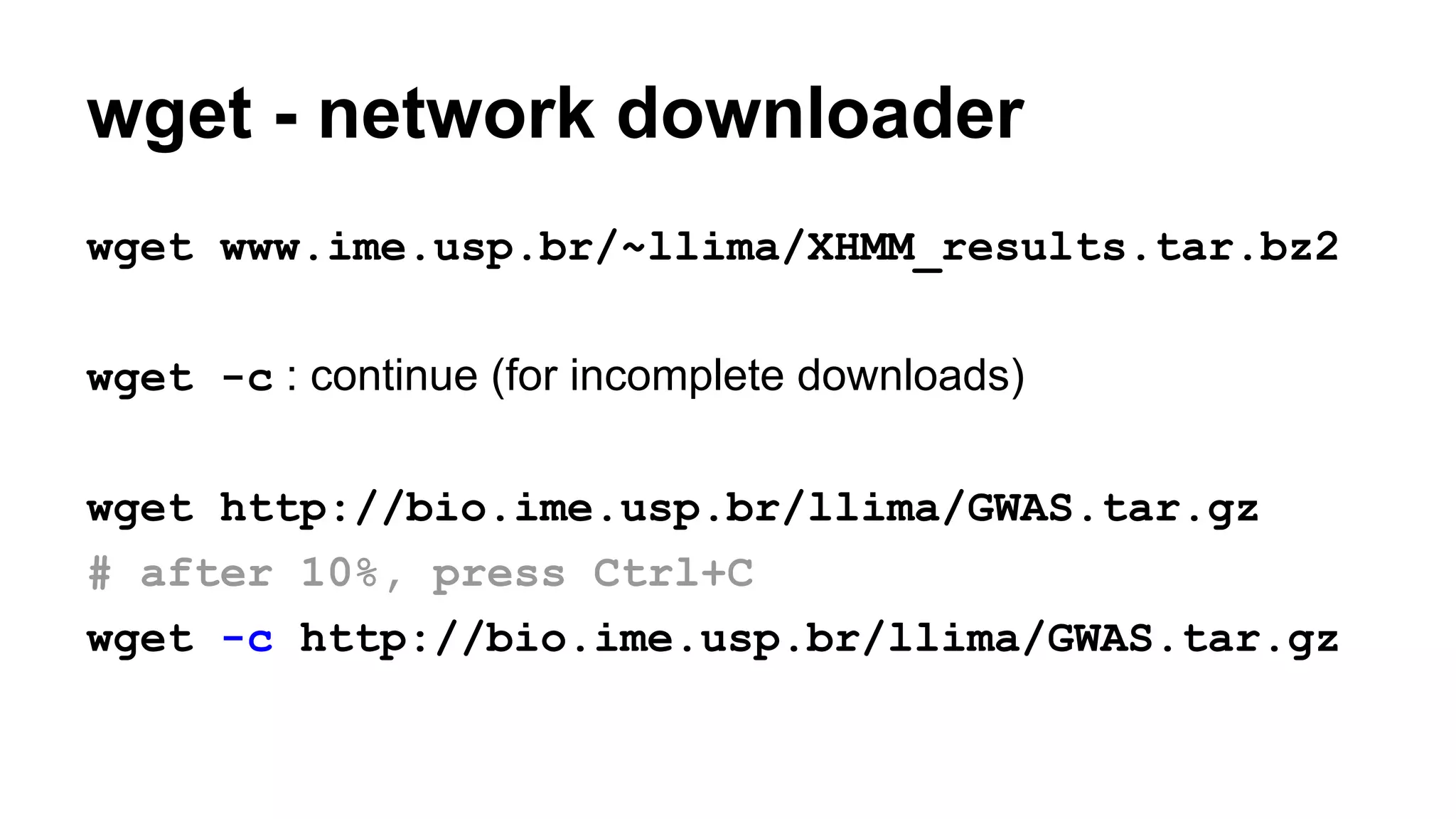
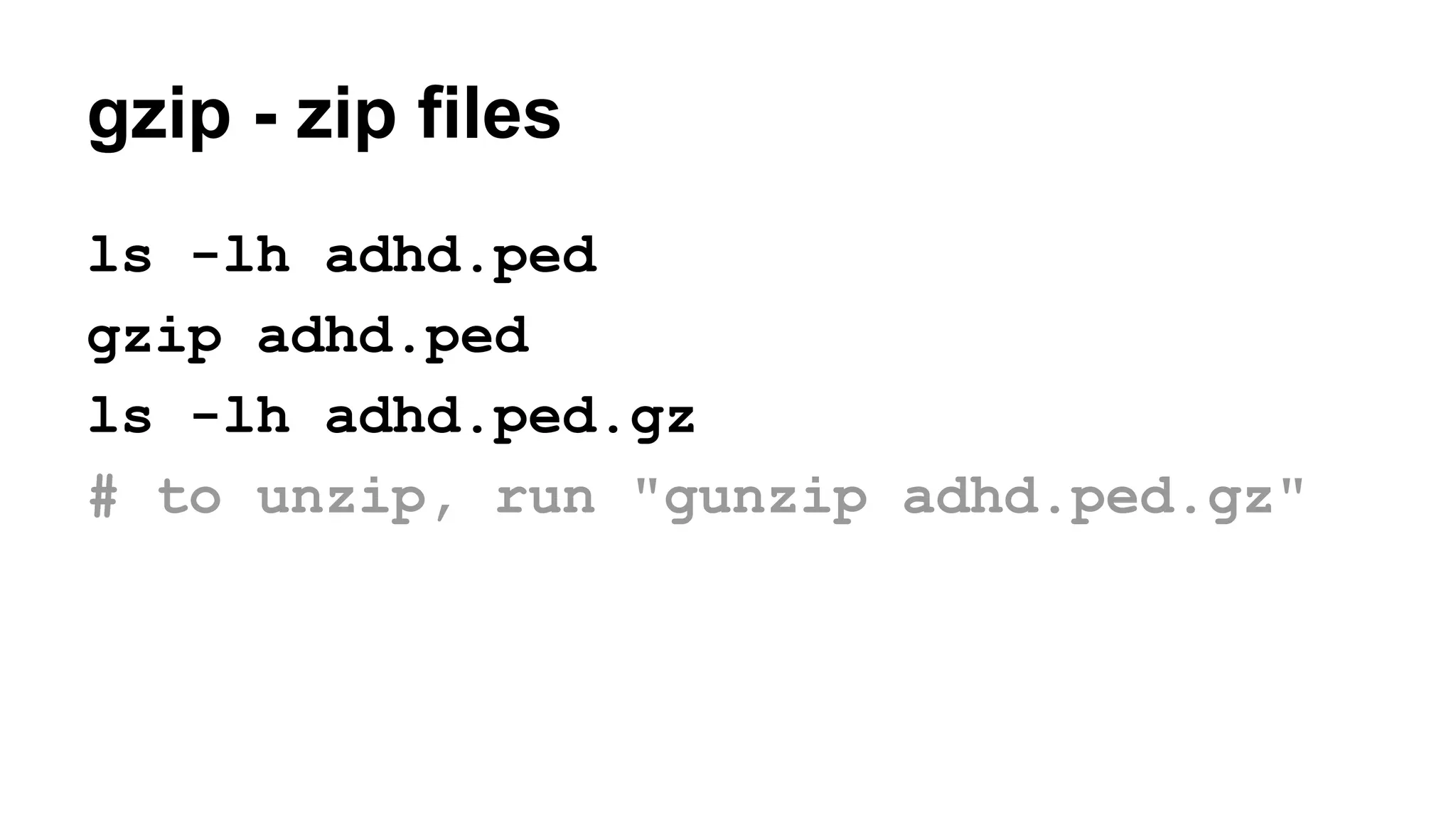
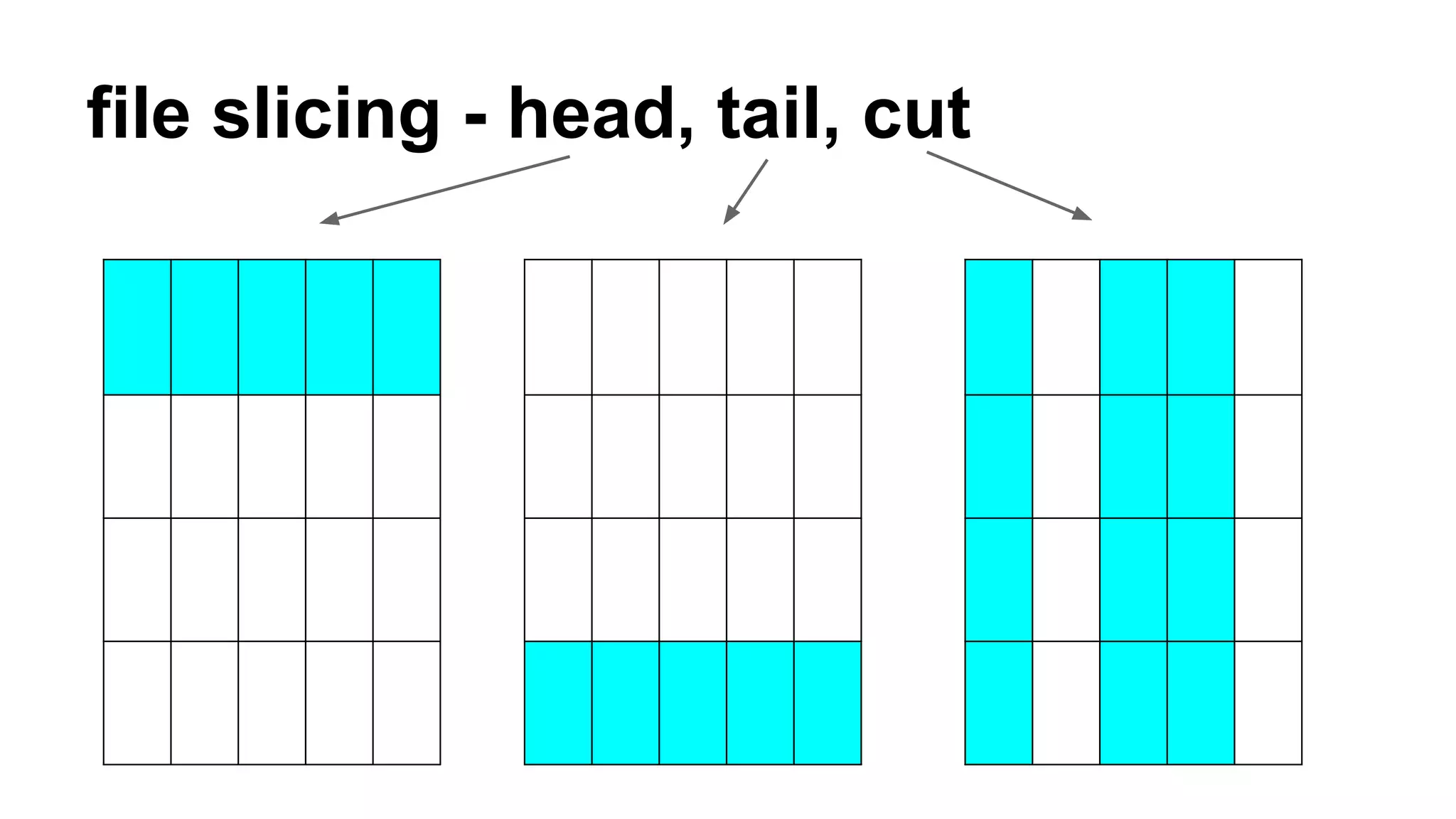
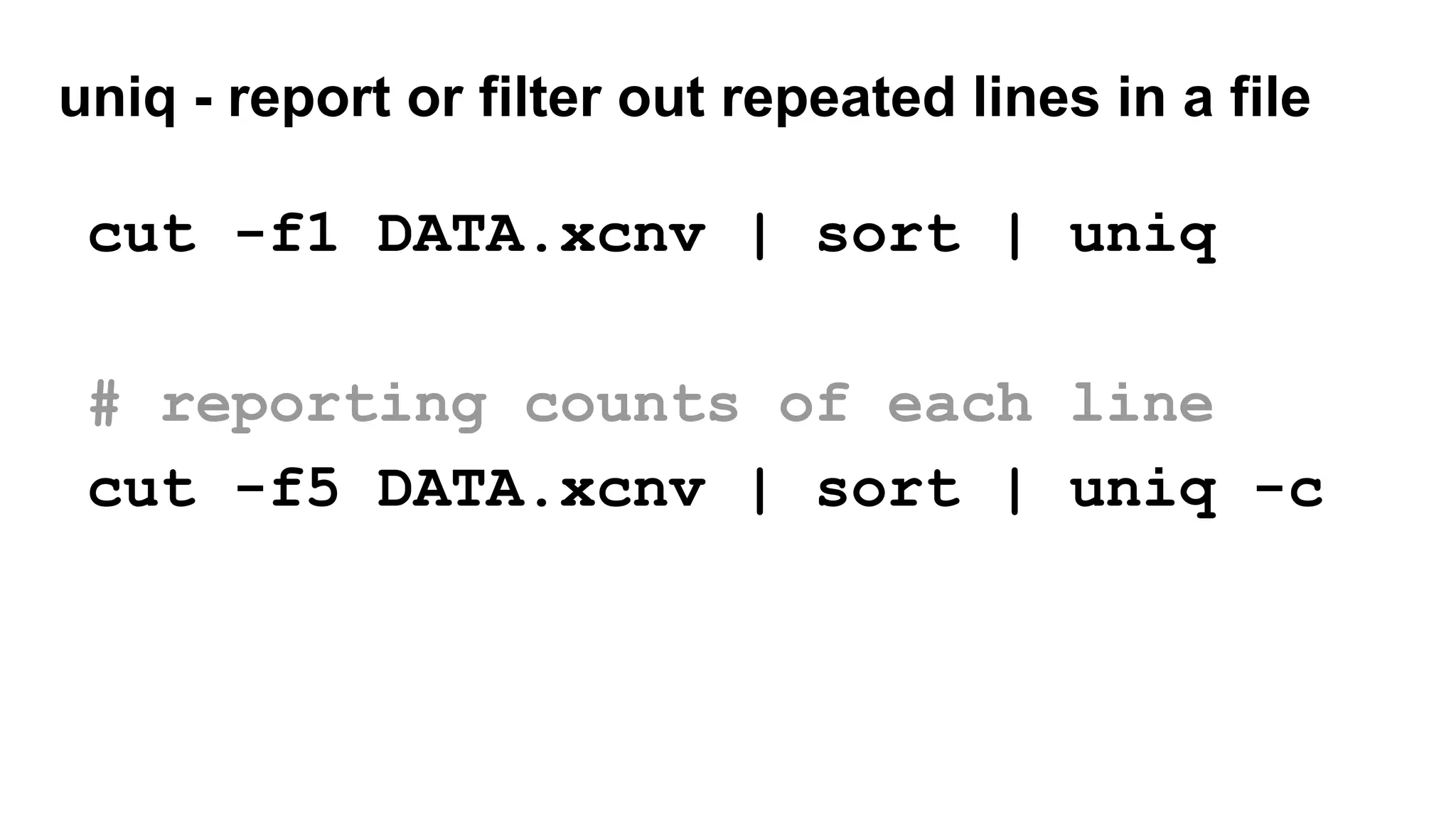
The document outlines a workshop on command-line tools, covering basic commands, file manipulation, text editing, and data processing techniques in a Linux environment. It provides examples of command usage, such as navigating directories, managing files, using special characters, and utilizing various commands for tasks like searching and sorting data. Tips for efficient command-line usage and exercises for practice are also included.






![Special characters (i)
^ : beginning of line
$ : end of line or beginning of variable name
? : any character (with one occurrence)
* : any character (with 0 or more occurrences)
# : start comments
[ ] : define sets of characters](https://image.slidesharecdn.com/workshopofcommand-linetools-day1-150213151151-conversion-gate02/75/Workshop-on-command-line-tools-day-1-7-2048.jpg)