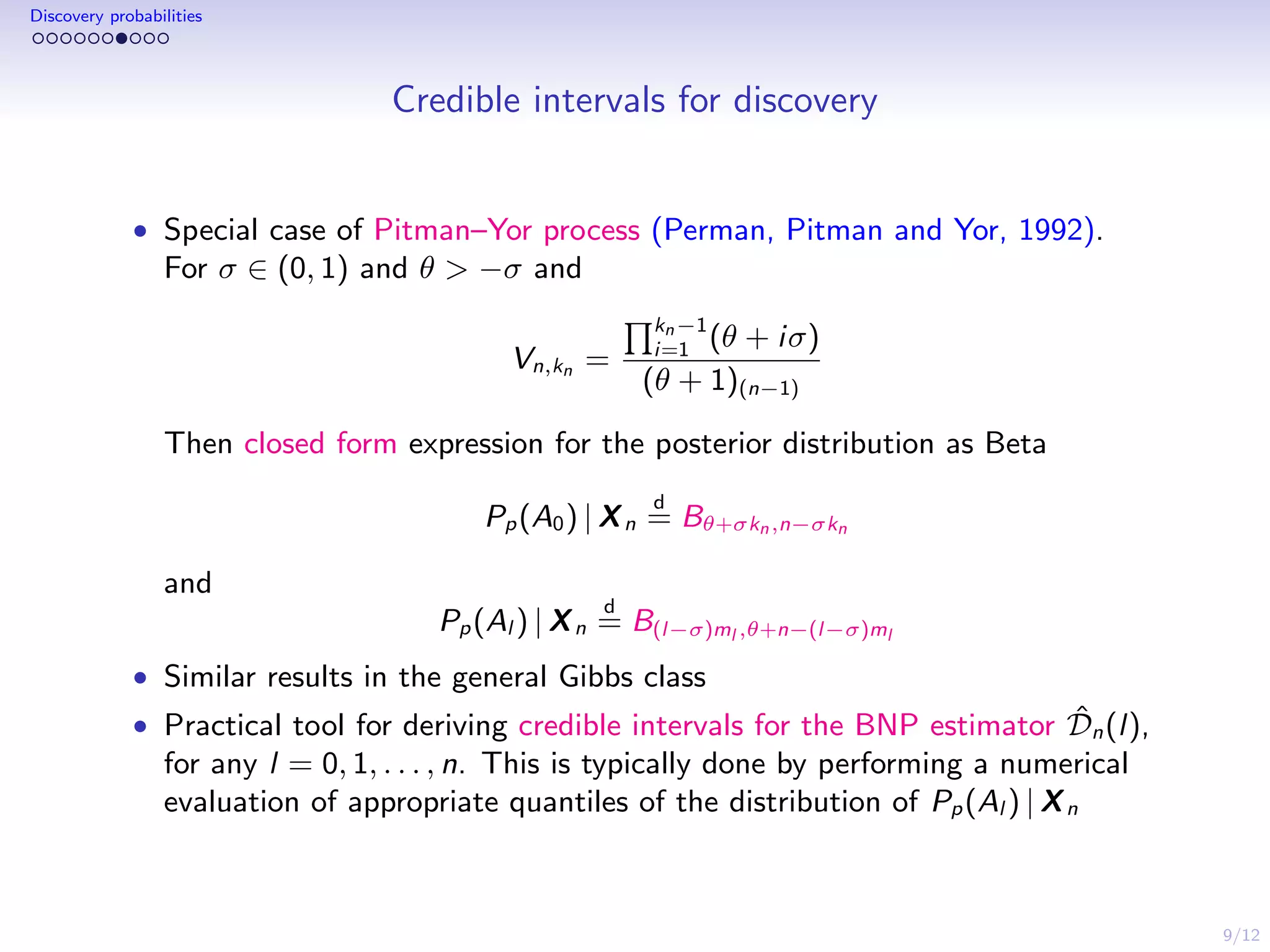
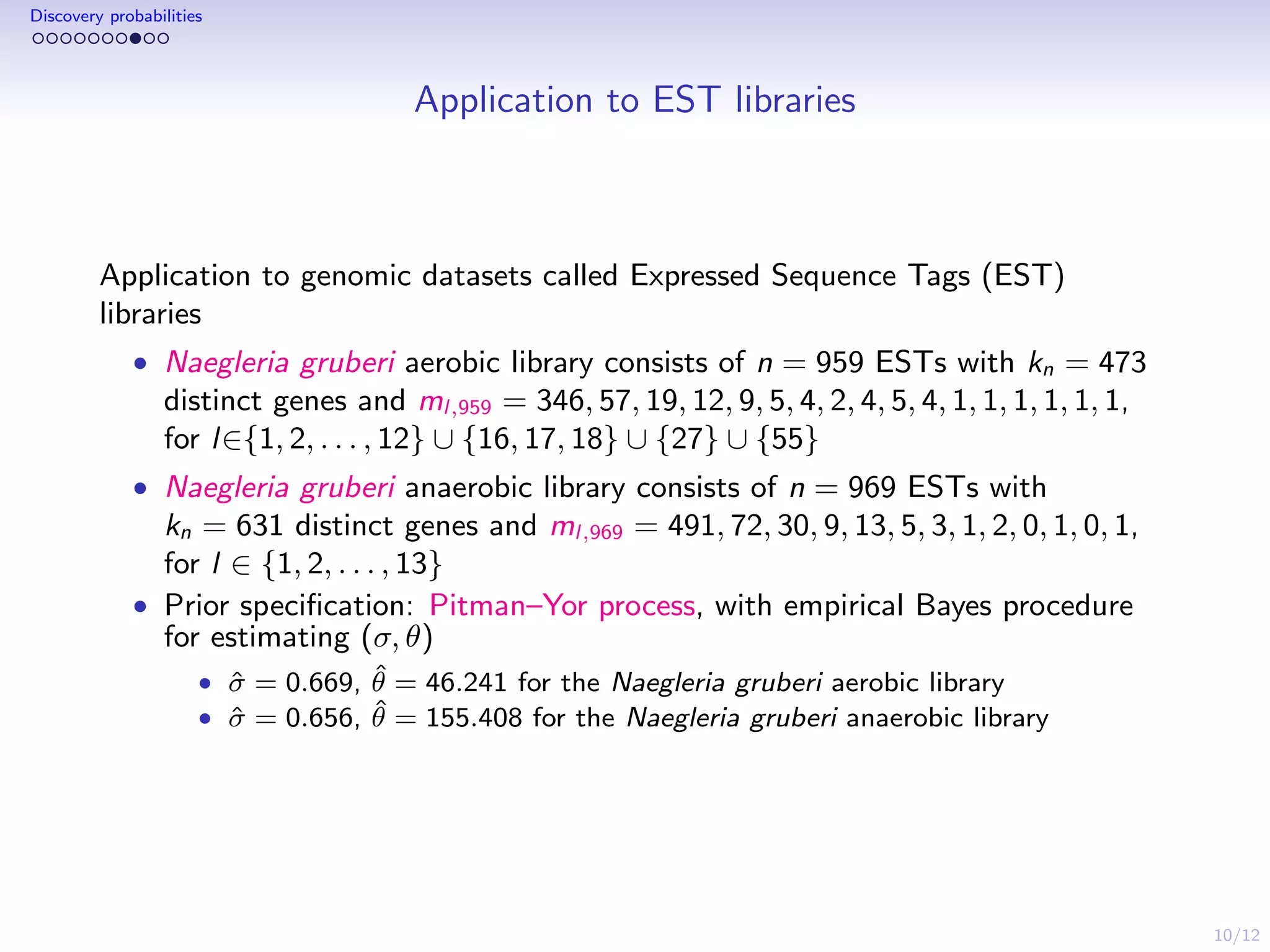
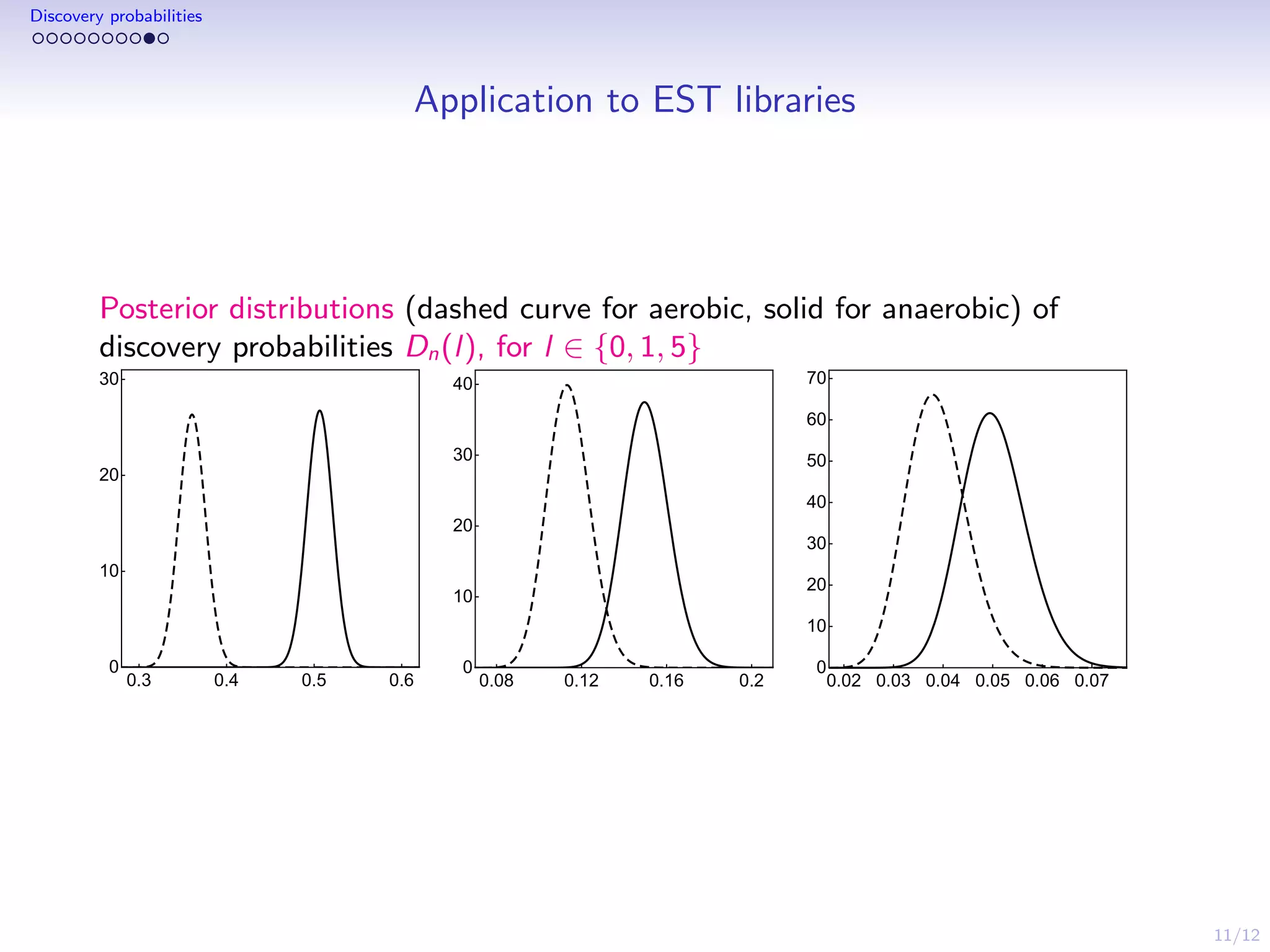
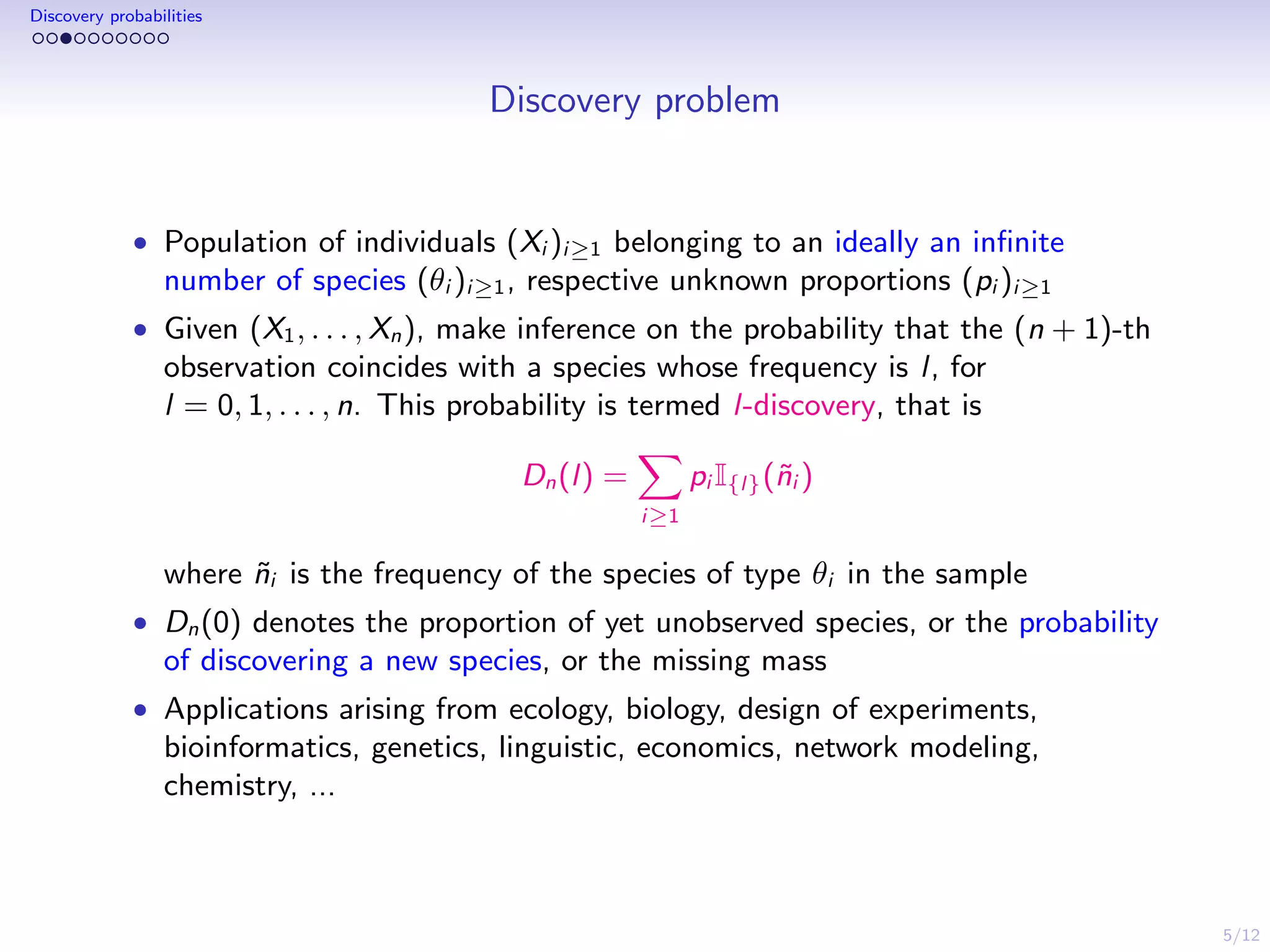
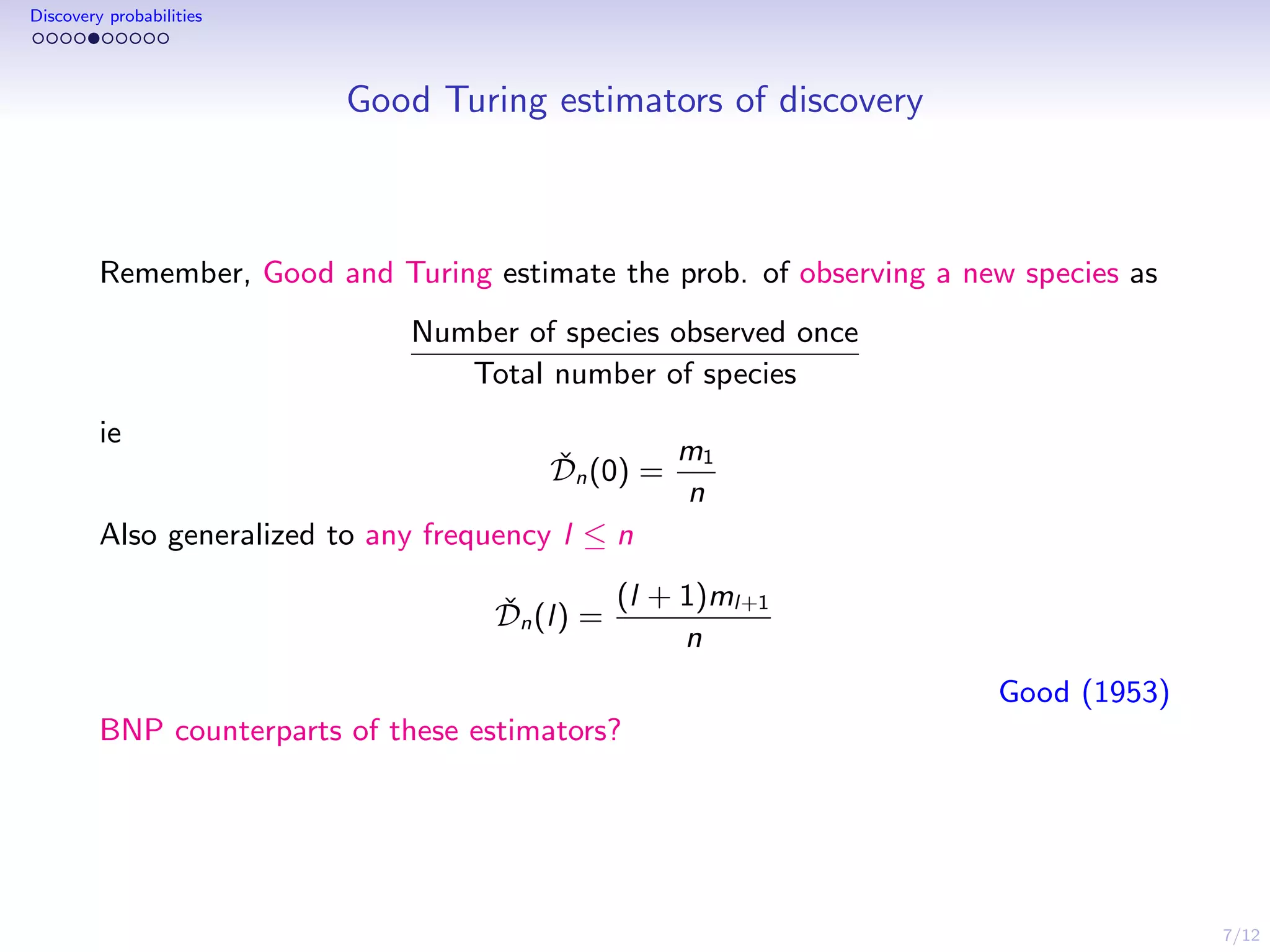
This document discusses species sampling models and discovery probabilities. It introduces the problem of estimating the probability of observing a new species given a sample. Good and Turing proposed an estimator for this during World War II. Bayesian nonparametric models provide an alternative approach by placing a prior on unknown species proportions. The document outlines BNP estimators for discovery probabilities and how credible intervals can be derived. It applies these methods to genomic datasets of expressed sequence tags to estimate discovery probabilities for observing new genes.




![8/12
Discovery probabilities
BNP estimators of discovery
Gibbs-type random probability measure P with index σ ∈ (0, 1): it is
characterized by (it induces) a predictive distribution of the form
P[Xn+1 ∈ A | Xn] =
Vn+1,kn+1
Vn,kn
G0(A) +
Vn+1,kn
Vn,kn
kn
i=1
(ni − σ) δX∗
i
(A),
BNP estimator ˆDn(l) of Dn(l) derived from the predictive using sets
A0 = X{X∗
1 , . . . , X∗
Kn
} and Al = {X∗
i : Ni,n = l}
BNP Good Turing
ˆDn(0) = E[Ph(A0) | Xn] =
Vn+1,kn+1
Vn,kn
ˇDn(l) = m1
n
ˆDn(l) = E[Ph(Al ) | Xn] = (l − σ)ml
Vn+1,kn
Vn,kn
ˇDn(l) =
(l+1)ml+1
n](https://image.slidesharecdn.com/statalksjulyanssm-160224162217/75/Species-sampling-models-in-Bayesian-Nonparametrics-8-2048.jpg)