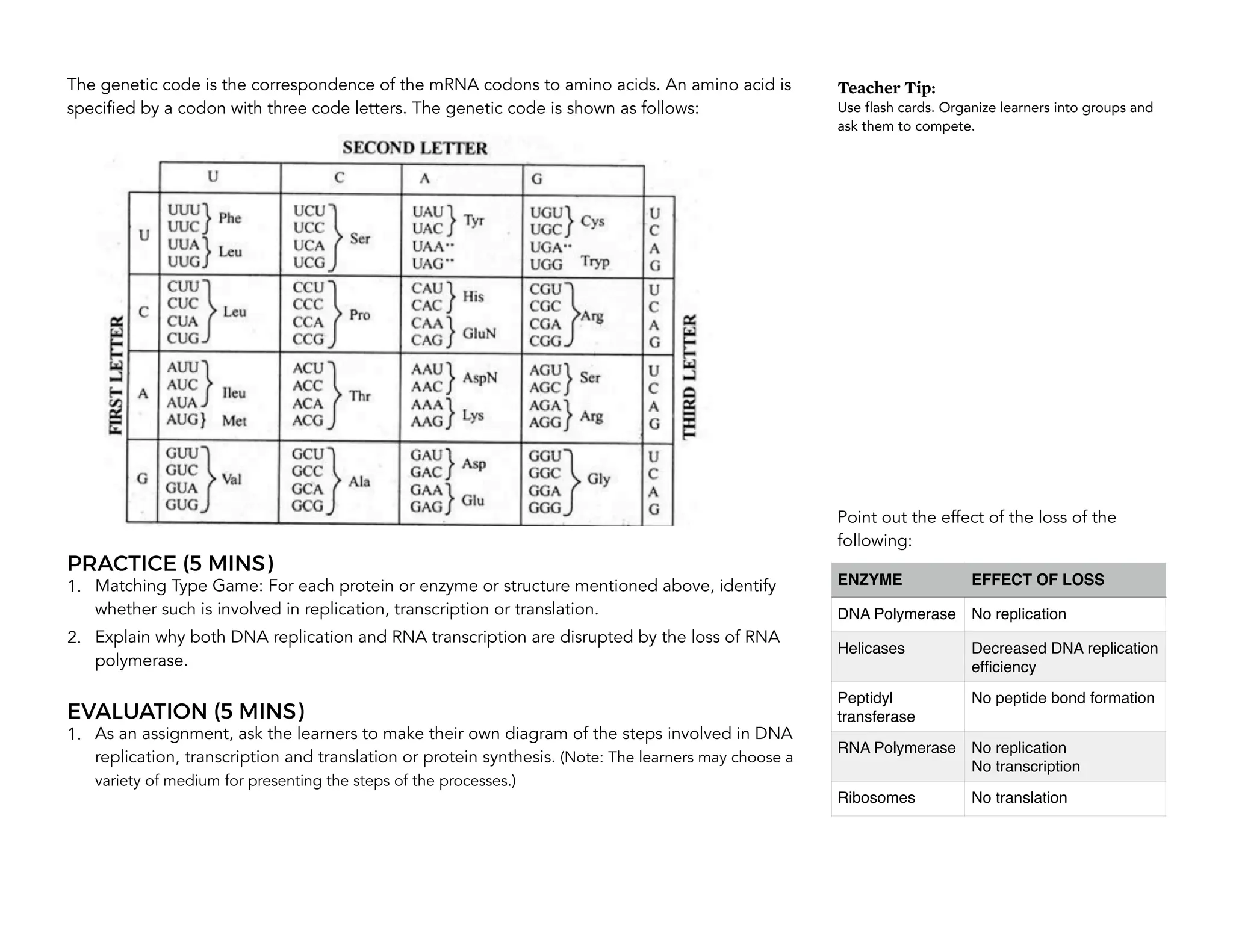
This document provides an introduction to the Teaching Guide for Senior High School General Biology 2. It was collaboratively developed by educators from various institutions to support the implementation of the K to 12 program. The guide is intended for STEM strand teachers and focuses on heredity and variation, diversity of living organisms, their structure, function, and evolution at a macro level. It introduces the SHS for SHS framework to promote meaning, mastery and ownership of learning. The framework aims to empower teachers as designers, facilitators and learners of their own lessons.