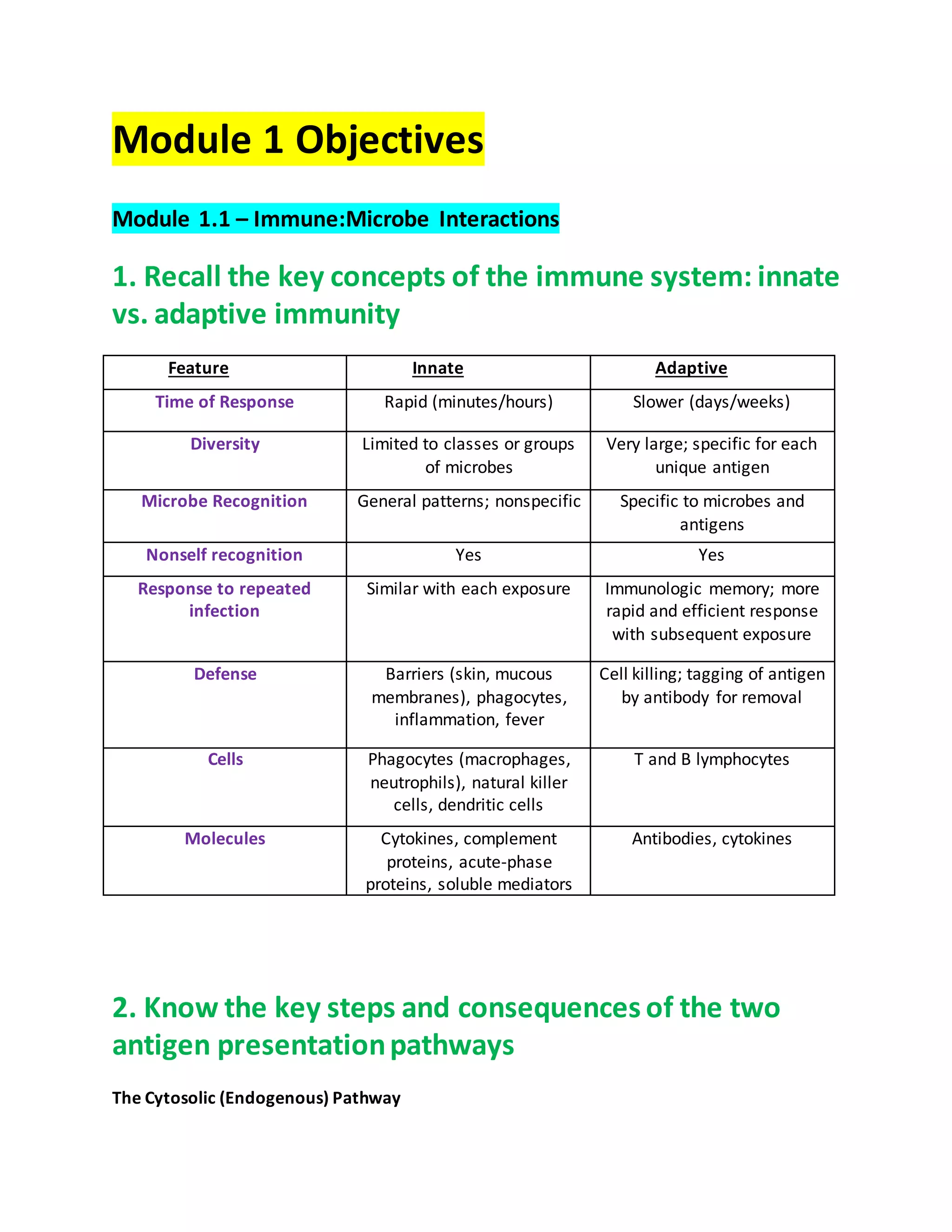
1. Module 1 covers immune-microbe interactions, antigen presentation pathways, and early life immune responses. It also covers defenses against extracellular and intracellular bacteria.
2. The adaptive immune system defends against intracellular bacteria through dendritic cell antigen presentation to CD4 T cells, which activate macrophages to kill bacteria via ROS/RNS. Intracellular bacteria enter host cells through mechanisms like T3SS injection and phagocytosis induction.
3. Intracellular bacteria evade immunity by preventing phagolysosome formation, inhibiting neutrophil free radical production, and escaping the phagosome into the cytosol. The adaptive immune system and inflammation contribute to defenses against extracellular bacteria through antibodies, cytokine production, and acute inflammation.




![o Secretary: membrane markers, enzymes, receptors, oxidases
Examples of neutrophil weaponry:
o Enzymes: lysozyme, NADPH oxidase complex
o Receptors: TLRs, complement receptors, Fc receptors, Formyl peptide receptor
(recognizes bacterial fMLP)
o Defensins: small proteins forming pores to lyse gram negative and gram-positive
bacteria, as well as fungi, yeast, and certain viruses.
o Lactoferrin: an iron chelator; makes iron unavailable for microbial growth
o Proteinases: MMP-8 and MMP-9 are matrix metalloproteinases that break down
collagen. Elastases break down elastin.
o Neutrophil free radicals: NADPH oxidase (comprised of p22, gp91 [membrane
bound] and p40, p47, and p67 [cytosolic] subunits) generates superoxide anions.
Superoxide anions can be converted to hydroxy radicals via SOD and Fe2+
via a Fenton reaction.
Cellular bleach (hypochlorous acid) is created from hydrogen peroxide
using free chloride ions and the myeloperoxidase enzyme.
These radicals attack nucleic acid, protein and lipid molecules to destroy
the ingested microbe.
There is a disease associated with the inability of neutrophils to produce superoxide
radicals called Chronic Granulomatous Disease (CGD)
o The disease is an inherited mutation effecting the NADPH oxidase subunits and is
X-linked, so mostly effects boys.
o These patients cannot fight off bacterial or fungal infections that are usually mild
or nothing to worry about in normal individuals. They are susceptible to
repeated infections.
3. Know how extracellular bacteria evade the immune
system
Bacterial evasion of skin and mucosal mechanisms and barriers:
Physical injury to skin
Mucosal bacteria can passively enter tissue
Mucosal bacteria can also actively invade deeper tissues through multiple mechanisms
including using enzymes to destroy tight junctions.
H. pylori produces urease to generate ammonia to neutralize stomach acid
Inflammation caused by initial bacterial infection could lead to tissue damage, allowing
bacterial entry.
Bacterial evasion while inside the body:](https://image.slidesharecdn.com/shaynemckeemedmicrostudyguide-210515001809/75/Medical-Microbiology-Study-Guide-by-Shayne-McKee-PharmD-2022-6-2048.jpg)




![2. Describe innate immunity against viral infections
Cytokines
Immune cells use TLR-3,7,8,9, and 13 to recognize viruses. TLR recognition activates
cytokine production in the immune cell
Viral infection causes almost all cell types to produce the type I cytokine IFN-1, mainly
coming from dendritic cells, and the type II cytokine IFN-gamma is produced from T and
NK cells.
o IFN-1 is highly potent against viruses. It can:
Reduce host cell viral receptor expression to reduce viral entry
Inhibit viral protein synthesis in the host cell
Activate macrophages and NK cells
Promote overall immune responses
The viral dsRNA can also be recognized by enzymes (PKR [protein kinase R], RNAse L-2-
5A-dependent ribonuclease) that can inhibit viral protein synthesis and induce cytokine
production.
NK Cells
Have KIR (Killing Inhibiting Receptors) CD159a, CD158,
CD94
Have KAR (Killing Activating Receptors) CD314
Have SHP-1: an SH2 domain-containing the phosphatase
1 protein (inactive when free, becomes an active
phosphatase enzyme when bound to KIRs)
Have DAP10: activates PI3 kinase when bound to the
CD314 KAR
Have perforin and granzyme that kill infected cells.
Notice the KIR, KARs, and
DAP10 are membrane bound](https://image.slidesharecdn.com/shaynemckeemedmicrostudyguide-210515001809/75/Medical-Microbiology-Study-Guide-by-Shayne-McKee-PharmD-2022-11-2048.jpg)










































![Bacillus Species Summary
All spp: saprophytic (needs only carbon and nitrogen for energy/growth), usually not
associated with human infections. All are gram positive rods, facultative anaerobes, and
acid-fast test negative.
The bacillus species virulence:
Spore forming, allowing withstanding of tough environments
Anti-phagocytic capsule
Bacillus anthracis
Gray to white, “ground glass” appearance
Hemolysis: gamma
Motility: non-motile
Anthracis specific virulence
o Toxins
Protective antigen (PA)
Allows binding to a cell and forms a membrane channel
Edema factor (EF)
Lethal factor (LF)
PA and EF combine to form edema toxin – causes cell and tissue edema
PA and LF combine to form lethal toxin to cause host cell death
PA and LF also impair innate and adaptive immunity and help with
bacteria proliferation
Infections/diseases/health risks:
o Cutaneous anthrax (95%)
o Inhalation [pulmonary] anthrax (5%)
o GI anthrax (rare)
o Biological warfare/bioterrorism
Anthrax most commonly seen in Asia/Africa
Treatment:
o Ciprofloxacin (fluoroquinolones) OR Doxycycline plus 1-2 of the following:
Rifampin
Vancomycin
Penicillin
Imipenem
Clindamycin
Clarithromycin
o Anthrax prophylaxis with short-lived vaccines](https://image.slidesharecdn.com/shaynemckeemedmicrostudyguide-210515001809/75/Medical-Microbiology-Study-Guide-by-Shayne-McKee-PharmD-2022-54-2048.jpg)















![o *When the bacteria are producing the ESBL enzyme, such as in some nosocomial
isolates, you can only use meropenem
Enterobacter, Citrobacter,and SerratiaSpecies
Summary (Enterobacter family)
Virulence factors (for all species)
All contain an inducible ampC gene that codes for beta-lactamases that can degrade
antibiotics, giving resistance to:
o Ampicillin
o Cefazolin
o Cephalexin
o Cefuroxime
Each can carry plasmids encoding resistance to multiple antibiotics (e.g. some may carry
plasmids for ESBLs, etc)
Treatment (for all species)
Cefepime
Meropenem
Ciprofloxacin
Enterobacter cloacae and Enterobacter aerogenes
Characteristics:
Gram negative bacilli, facultative anaerobes
Lactose fermentation positive
Oxidase test negative
Hemolysis negative
Voges-Proskauer reaction positive
Motile
Mucoid growth (colonies are sticky/viscous)
Normally found in the GI
Infections:
Mostly associated with respiratory tract infections (pneumonia)
UTIs (with indwelling [inside the body] catheter use)
Intra-abdominal infections
Wounds or burns – skin infections](https://image.slidesharecdn.com/shaynemckeemedmicrostudyguide-210515001809/75/Medical-Microbiology-Study-Guide-by-Shayne-McKee-PharmD-2022-70-2048.jpg)
![Citrobacter koseri and Citrobacter freundii
Characteristics:
Can use citrate as their only carbon source
Produce H2S
Gram negative, facultative anaerobes
Lactose fermentation positive
Oxidase test negative
Voges-Proskauer reaction negative
Commonly found in GI tract of animals and humans
Commonly found in soil and water
Infections:
Mostly associated with UTI’s (with indwelling [inside the body] catheter use)
Respiratory tract infections
Intra-abdominal infections
Wound infections
Osteomyelitis
Serratia marcesens
Characteristics:
Widespread in the environment due to its saprophytic characteristics, although not
common in human fecal flora
Produces the exotoxin DNase
Produces a red pigment – prodigosin
Gram negative bacilli, facultative anaerobe
Lactose fermentation positive
Oxidase test negative
Hemolysis negative
Voges-Proskauer reaction positive
Normal flora, but mostly found in the environment
Serratia-specific Virulence:
Contain fimbriae that allow adherence to uroepithelial cells and can be cytotoxic to
some the tissue cells
Can survive harsh conditions, even disinfectants
Its ampC gene produces higher levels of beta-lactamases during antibiotic therapy](https://image.slidesharecdn.com/shaynemckeemedmicrostudyguide-210515001809/75/Medical-Microbiology-Study-Guide-by-Shayne-McKee-PharmD-2022-71-2048.jpg)
![Infections:
Mostly associated with UTIs (with indwelling [inside the body] catheter use)
Respiratory tract infections
Wound infections
Osteomyelitis
Intra-abdominal infections
Other Important Enterobacteriaceae Summary
(Enterobacter family)
Salmonella
There are two species
Salmonella bongori
Salmonella enterica
o Subspecies:
Salmonella enterica salamae
Salmonella enterica arizonae
Salmonella enterica diarizonae
Salmonella enterica houtenae
Salmonella enterica indica
Salmonella enterica enterica
Sub-subspecies (serotypes) that cause enteric fever:
o S. paratyphi A (serogroup A)
o S. paratyphi B (serogroup B)
o S. choleraesuis (serogroup C1)
o S. typhi (serogroup D)
Infections from Salmonella:
o Overview:
Usually cultures will be from stool or blood. Hospitals will be able to
determine if Salmonella is present, but to identify the serotype, the
sample is sent to a state or regional reference lab.
Usually the four serotypes enter the body orally with contaminated food
or drinks.
Our body’s protective factors:
Gastric acidity
Normal intestinal microbiota
Local intestinal immunity](https://image.slidesharecdn.com/shaynemckeemedmicrostudyguide-210515001809/75/Medical-Microbiology-Study-Guide-by-Shayne-McKee-PharmD-2022-72-2048.jpg)














![NeisseriaeSpecies Summary
Characteristics and general facts:
Gram-negative diplococci
Grow in Thayer-Martin (chocolate [lysed] sheep blood and antibiotics)
o Growth is slow and performs better in CO2 rich environments
Colonies appear translucent, nonpigmented, and glisten (mucoid
growth) and are hemolysis negative. Colonies are also oxidase positive
(turns purple)
Quick comparison between species (same information is presented below in list format)](https://image.slidesharecdn.com/shaynemckeemedmicrostudyguide-210515001809/75/Medical-Microbiology-Study-Guide-by-Shayne-McKee-PharmD-2022-87-2048.jpg)










![ Invasive devices
Localized sources of infection (e.g. pyelonephritis)
o Symptoms of cutaneous infection:
Thrush
Vulvovaginal candidiasis
Localized skin reactions
o Symptoms of systemic infection:
Candidemia
Infection of internal organs like kidneys, heart, brain
Candida may infect hardware (e.g. prosthetic heart valves)
Treatment options and theirresistance mechanisms
o Triazoles (fluconazole)
Efflux out of yeast cells
Altered drug binding site
Alternative ergosterol pathways
o Polyenes (amphotericin B or nystatin)
Alternative ergosterol pathways or low ergosterol levels in the cell wall
o Echinocandins (micafungin)
Altered binding to target site
Haemophilus Influenzae Summary
HaemophilusInfluenzae
Characteristics
o Gram-negative, facultative coccobacillus, pleomorphic (morphology
changes with length of incubation)
o Considered normal respiratory flora
o Grows best on chocolate agar (lysed blood) and in the presence of CO2
Requires presence of X (hemin) and V (nicotinamide adenine
dinucleotide [NAD]) factors for growth which are contained in
the chocolate agar
o Colonies appear large, flag and colorless (or gray)
o Non-hemolytic
o Odor
o Encapsulated forms appear mucoid
o Can be found as a satellite organismfor pathogens that lyse RBC’s (e.g.
staph aureus) This means that if staph aureus is growing on a plate and
breaking down RBC’s within the chocolate agar, haemophilus may be](https://image.slidesharecdn.com/shaynemckeemedmicrostudyguide-210515001809/75/Medical-Microbiology-Study-Guide-by-Shayne-McKee-PharmD-2022-98-2048.jpg)

















