This document reports on a laboratory project investigating the role of the non-coding RNA rli60 in Listeria monocytogenes. The student constructed an rli60 knockout strain of L. monocytogenes and examined gene expression and metabolism compared to the wild type strain when grown in different media. Quantitative PCR results showed that rli60 is involved in down-regulating the ilv operon, which encodes for branched amino acid biosynthesis. Growth curve and luminescence assays indicated normal bacterial growth and hly virulence gene expression in the rli60 mutant strain. In summary, this project found that the non-coding RNA rli60 regulates metabolic gene expression but does not affect growth or a key virulence factor in L. monocytogenes

![1
Abstract
The genes involved in bacterial pathogens adaptations to their host are under tight
regulation. Recently, non-coding RNAs (ncRNA) have emerged as key regulators of
such systems. Since the systems described here control, among others, the
biosynthesis of specific molecules which are vital for the pathogen during infection, it
is crucial to study them.
Science is always seeking for a new 'move', for an un-expected manipulation that will
raise an advantage in order to be one step ahead pathogens. The idea is a bit
pretentious, but is the same also in this basic project- trying to investigate whether the
ncRNA rli60 involves in such regulation, by generating a rli60 knock-out strain,
grown the mutant in different media, examining gene expression and searching for
affects over metabolism and virulence in comparison to Listeria monocytogenes wild
type (WT) strain. Here I show that rli60 is involved with the down-regulation of a
main metabolic operon- ilv. This conclusion makes room for a better understanding
over L.monocytogenes life cycle and perhaps will serve as a basic pioneer way of
dealing with listeriosis.
Introduction
L.monocytogenes is a gram-positive, non-sporulating, rod shaped, facultative
anaerobe that is a member of the phylum firimicutes and the causing agent of
Listeriosis. Listeriosis is an uncommon but very serious condition that has a high
mortality rate among susceptible individuals and is acquired orally through the
consumption of spoiled foods [1].
In recent years, L.monocytogenes has emerged as a model organism in infection
biology and also has become an attractive system for the study of gene regulation and
especially regulatory RNAs in pathogenic bacteria [4].
The concept of nucleic acids being involved in gene regulation is relatively new. Not
long ago, it was accepted that only proteins control gene regulation. This dogma does
make sense due to the complexity of the diverse regulatory systems, which can be
solved probably only by the complicated structure and numerous different kinds of
proteins. On the other hand, this dogma can also be easily contradicted by the
which holds that life originally existed,hypothesis"RNA world"n ancientcommo
later.alongoteins cameprcomplex amino acid structures suchly RNA, andusing on
bealsocouldnctions performed by proteinshis hypothesis requires that all critical fuT
.[1]independentlyperformed by RNA
he firstTgroups.three mains intobacterial regulatory RNAsifyWe can generally clas
are elements present in the 5 UTR of the mRNA that they regulate (e.g: riboswitches,
thermosensors and pH sensors). The second, trans-encoded small RNAs (sRNAs),
which are defined as regulators of one or several target genes located elsewhere on the
chromosome. And the third, cis-encoded antisense RNAs (asRNAs) that overlap and
are complementary to their target genes encoded on the opposite DNA strand of the
same genomic locus [2].](https://image.slidesharecdn.com/a18316e7-86e9-49b9-a490-677ac65927dc-161006113259/85/rli60-project-FINAL-2-320.jpg)
![2
In this project I focused on a non-coding RNA (rli60) in order to investigate its
role as regulatory RNA. This sequence is localized up-stream the ilv operon
which is regulated by CodY in L.monocytogenes.
, among others,s, the first group of bacterial regulatory RNAs consists mentionedA
because ofriboswitchfunction asrli60It is tempting to speculate thatriboswitches.
few supportive reasons.
the fact thatnda,all domains of lifeinnce of riboswitcheshe existetFirst,
are an effective method of controlling gene expression in naturalriboswitches
.[5]organisms
thealsoand,[2]in a long intergenic regionfoundisrli60In addition, the fact that
)1(figure 1rli60ofat the 3'palindromesC reach-Gand twotailU-polyaexistence of
-downwithwhich can interfereindependent terminator-Rhofunction asthat can
transcription, contributes to the assumption above.stream genes
can bind small moleculessRNARiboswitches demonstrate that naturally occurring
a capability that many previously believed was the-as mentioned earlierspecifically,
It has been.ptamersAor artificially constructed RNAs calledproteinsdomain of
ry systems, or evensuggested that some riboswitches might represent ancient regulato
conservedgenerallydomains arewhose bindingribozymesworld-RNAremnants of
and anptamerARiboswitches are often conceptually divided into two parts: an[3].
expression Platform [6]. The Aptamer directly binds the small molecule, and the
expression Platform undergoes structural changes in response to the changes in the
Aptamer. The expression Platform is the component which regulates gene expression.
Figure 1. A general sketch of a riboswitch. The binding of a metabolite leads to a structural changes at the expression Platform
which then regulates gene expression [6].
Expression platforms typically turn off gene expression in response to the small
molecule, but some activate it. According to previous studies, riboswitches can
regulate gene expression through transcription- by controlling the formation of rho-
independent transcription termination hairpins which can lead to premature
transcription termination- and also through translation- by mediating some folding
that sequesters the ribosome binding site (RBS) and thereby inhibiting translation.](https://image.slidesharecdn.com/a18316e7-86e9-49b9-a490-677ac65927dc-161006113259/85/rli60-project-FINAL-3-320.jpg)
![3
Furthermore, the riboswitch can function as a ribozyme that cleaves itself in the
presence of sufficient concentrations of its metabolite and also can alternate structures
that can affect the splicing of the pre-mRNA [6].
The ilv operon encodes for the biosynthesis of the branched amino acids (BCAA)-
Isoleucine, Leucine and Valine. It is redundant to say that these amino acids are
critical for L.monocytogenes growth and especially in matter of intracellular infection
where the pathogen must survive low cytosolic concentrations of such amino acids in
the host cell since mammalian cells do not produce them endogenously. In addition,
low concentration of BCAAs leads to elevated transcription of virulence genes [7].
Therefore, the study of the regulatory system of the ilv operon and the transcription
through infection is crucial for the understanding of such complex mechanism.
thodseand MsMaterial
:rli60-deletion mutantonstruction of aC
The whole work will be follow by this sketch:
Fig 2. Sketch of the rli60 locus in L.monocytogenes genome. This drawing shows the first step in the rli60 mutants'
construction: the direction and purposes of the primers is given.
primers A, B, C and Dthegnand constructidesigning:STAGE1
i. The primers sequences:
CAC ATC ATC ACT CTT CCT TGAT TCCG GG AGC TCGAC ATG ATT ACG AAT TC-(primer A) '5
(primer B)'5-CAA AAGATT GTA AAG AAC TAT AAT TAA GCTCG TTG GTA TAT ATA ATT TAT GAT TGT
AAG CAT CGA AAA GCTAA CAT TTC TTG ATA TTA ATT CGA GTT TTC-5'(primer C)
(prime D)5'-CTA GGA GAT CTCGGG CCCC ATA ACT TCT GAT GCT AAA CCT TGC GAT
A sense
B anti
C sense
D anti
C anti B sense
~1000 bp
ATG TAA
C
rli60
~800 bp
B
Xma1Sac1](https://image.slidesharecdn.com/a18316e7-86e9-49b9-a490-677ac65927dc-161006113259/85/rli60-project-FINAL-4-320.jpg)



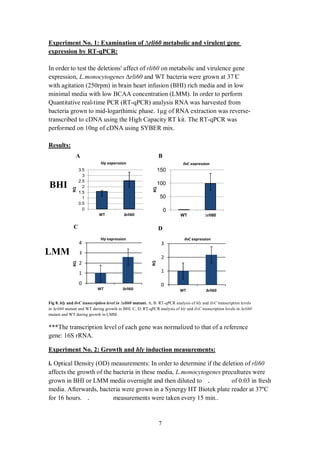
![8
Results:
ii. Relative luminescence measurements (RLU): for luminescence assays a plasmid
harboring the lux reporter system (pPL2- ) fused to the hly promoter and was used
In order to study the affect of the deletion of rli60 on hly transcription. pPL2-P
were conjugated to WT and to rli60 strains down-stream the hly promoter (see
drawing in fig. 9 ii) in a way that once the hly promoter is active we can measure
luminescence. Conjugated precultures were grown in LMM media overnight and then
diluted to . of 0.03 in fresh media. Afterwards, bacteria were grown in a
Synergy HT Biotek plate reader at 37ºC for 16 hours. luminescence measurements
were taken every 15 min.
Results:
Fig 9. Normal bacterial growth and normal transcription of hly in rli60. i. Optical density measurements of WT and rli60
L.monocytogenes cultures in BHI and in LMM. ii. Relative luminescence measurements (RLU) indicating activation of hly
promoter (Phly) under growth of WT and rli60 L.monocytogenes (harboring pPL2- plasmid) in LMM.
Discussion
This project objective was to examine the regulatory role, if any, of the ncRNA rli60
over metabolism and virulence of L.monocytogenes. The genes that were chosen in
order to represent those two categories were hly for virulence and ilvC for
metabolism.
Upon cell entry, L.monocytogenes escapes from the phagosome/vacuole into the host
cytosol by producing the pore-forming hemolysin toxin, listeriolysin O (LLO) which
encoded by the hly gene [7]. Thus, during intracellular infection there is an increase of
0
0.15
0.3
0.45
0.6
0.75
0 4 8 12
WT (BHI)
rli60 (BHI)
WT (LMM)
rli60 (LMM)
O.D600nm
Time (h)
0.E+00
5.E+04
1.E+05
2.E+05
2.E+05
3.E+05
0 6 12 18 24
WT (LMM)
rli60 (LMM)
Time (h)
RLU
luxPhly](https://image.slidesharecdn.com/a18316e7-86e9-49b9-a490-677ac65927dc-161006113259/85/rli60-project-FINAL-9-320.jpg)
![9
hly transcription. This is the main reason for executing experiment presented in figure
9 (ii) in LMM- in order to resemble intracellular condition. In addition, in such case
L.monocytogenes metabolism demands BCAAs. ilvC is a part of the ilv operon and
encodes for the biosynthesis of ketol-acid reductoisomerase that is involve in BCAAs
biosynthesis [7], so also in this case I expect high transcription levels. On the other
hand, BHI is a rich media for L.monocytogenes growth so I can easily claim that in
this media WT strain will not increase the transcription of hly (since the conditions
that the media provides are different than the intracellular conditions that triggers hly
expression) and also will not increase transcription of ilvC because the bacteria
doesn't need to perform any special adjustments (e.g. biosynthesis of BCAAs) in
order to fit the environment. To summarize, during L.monocytogenes gowth in LMM
I expect to witness increase of hly and ilvC transcription in comparison to growth in
BHI. The results support this assumption partially (fig. 8). ilvC transcription in
different media fulfills the hypothesis above, but hly transcription levels doesn't- hly
transcription of WT in BHI is slightly higher than WT in LMM. Since the small
differences (1.5 RQ units vs. 1 RQ units) I can attribute this deviation to technical
errors. Experiment repeats are needed.
ilvC transcription level of rli60 mutant in BHI is 100 times higher than in WT
(figure 8 B). Hence, I can infer that rli60 is involved with down-regulation of ilvC.
The fact that the ncRNA of rli60 locus is up-stream to the ilv operon leads me to
speculate that maybe indeed rli60 functions as a riboswitch that regulates ilvC.
By examining the growth over time (fig 9 i) there is a predictable difference of growth
in BHI as opposed to LMM- enriched and faster growth in BHI, but there isn't any
major differences at all in the growth of rli60 in comparison to the WT strain. This
outcome can be refer to the assumption that although the deletion that I generated
( rli60) indeed affects ilvC transcription, the bacteria probably has found another way
to survive (e.g. another locus of sequence similar to the one that been deleted or
another way for producing BCAAs) resulting in a backup mechanism that will help to
overcome this manipulation
In addition, in spite of the expectation to see elevated growth in rli60 since rli60
negatively regulates ilvC and it should constitutively expressed now, there is no
change in the growth of rli60 in LMM in comparison to WT. The reason of this
outcome can be explained by another secondary regulation component that cover the
lack of rli60 and balance the ilvC levels resulting with naturally inhibition of the
growth.
Due to the results of Ex. 1 and 2 (fig. 8, 9) there is a small change in hly transcription
in the rli60 strain compared to the WT strain. The mutant shows a slightly higher
level of hly transcription in the WT and it is pretty hard to determine that rli60 has
also a negative regulatory role in hly transcription. Moreover, since rli60 locus is
~2000bp up-stream hly locus [8] I didn't suppose initially that rli60 functions as
direct riboswitch also in the hly case. Performing this experiment was intended to](https://image.slidesharecdn.com/a18316e7-86e9-49b9-a490-677ac65927dc-161006113259/85/rli60-project-FINAL-10-320.jpg)
![10
find out whether the deletion of rli60 will raise a change in the virulence of the mutant
that can may be explained by a 'third party' implication over hly transcription that
have been lost due to the deletion.
In order to claim better and more significant conclusion about rli60 and hly
relationship further studies must be executed.
esearchurther RF
The main result of this project infers that rli60 negatively regulates ilvC transcription.
By previous knowledge of this specific sequence, the initial suspicion was that rli60 is
a riboswitch who regulates this gene. The findings of this work contribute to this
speculation but of course- do not confirm it.
In order to understand whether rli60 functions as a major regulator of ilvC
transcription I can execute an experiment that will test rli60 ilvC transcription level
over increasing amounts of BCAAs in the media in comparison to the WTs'
transcription. As for the WT strain my expectation is to detect a decrease in ilvC
transcription since the media becomes richer in BCAAs. If rli60 is really a major
regulator of ilvC, I will receive a constant high level of ilvC transcription over
increasing amounts of BCAAs in the mutant rli60.
Over macro observation, success in proving that rli60 function as the major regulator
of ilvC transcription, can lead to a new opportunity dealing with L.monocytogenes
infection. If I can produce a constitutive mutation of rli60 that will block consistently
ilvC transcription it will interfere with the bacterias' ability to produce BCAAs and
then decrease its virulence.
Thus, if I had to continue this study, in order to construct such strain I must first
clarify if rli60 functions as a classic riboswitch. As mentioned, riboswitches consist of
two major domains- Aptamer and Expression Platform. In general, the metabolite that
binds the Aptamer is the end product of the pathway that it regulates [9]. Meaning that
there is a good chance that rli60 binds one of the BCAAs, for instance- Isoluecine. In
order to exam if RNA sequence binds a specific metabolite a simple and classic
experiment can be executed using RNase-T [9]. I can synthesis in-vitro rli60
sequence and radiolabel it. Then I can incubate the RNA with RNase-T in the
absence of the metabolite- Isoluecine, separate and run the spontaneously cleavage
products in gel-electrophoresis. Simultaneously, I will do the same but now adding
Isoluecine. If rli60 does bind Isoleucine I will receive an altered pattern of cleavage
products in gel-electrophoresis which indicates for the location of the binding site.](https://image.slidesharecdn.com/a18316e7-86e9-49b9-a490-677ac65927dc-161006113259/85/rli60-project-FINAL-11-320.jpg)
![11
Fig 10. RNA can bind metabolite. An Example of experiment that can be performed in order to show binding of a specific
metabolite to a RNA sequence. The location of metabolite X in RNA binding site has altered the picture and represent by the red
bracket.
In case that rli60 really does bind one of the BCAAs, I can try to predict the
secondary structure of rli60 [11] in order to have a better understanding of the
Aptamer's binding site, which might give the ability to manipulate that site and
construct a consistent mutation that will affect rli60 to repress consistently ilvC
transcription. Then I can measure the growth of the new mutant in comparison to WT
and to be convinced with rli60 regulatory importance.
In another direction, I can test the relationship between CodY and rli60. In
L.monocytogenes the protein CodY represses genes involved in amino acid
metabolism, nitrogen assimilation and sugar uptake in the presence of BCAAs, and is
important for the activation of virulence and metabolic genes necessary for
intracellular growth in the absence of BCAAs [7]. rli60 might consist a binding site
for CodY [7] (fig. 11) and perhaps this is the mechanism that originally regulates ilvC
transcription, meaning that the deletion of rli60 cause miss-regulation by CodY and
that is what led to high ilvC transcription level of rli60 (fig. 8). In order to define
whether rli60 regulate ilvC depending on CodY or as an independent riboswitch I can
construct a deletion mutant lacking only the CodY binding site in the rli60 sequence
and see if the regulation ability has been lost.
Dealing with affects of rli60 on hly, this project results can't give significant data that
will help to manage any rational conclusions. hly examining should be tested again.
For instance, due to the long distance between rli60 and hly I can search over
L.monocytogenes genome for compliment sequences to rli60 and test their gapping
with other critical genes that may relates with hly- perhaps rli60 function also as
trans-asRNA mediated gene regulation and affect their expression by attaching them
when needed and triggering the CRISPR complex [12].](https://image.slidesharecdn.com/a18316e7-86e9-49b9-a490-677ac65927dc-161006113259/85/rli60-project-FINAL-12-320.jpg)
![12
Supporting information
CodY box 6 mm CodY box 6 mm
TTTGACCAAACTATT CTGACTATAT ACTAAa acTaTaAAAA TaCAAATTaatTAA AtAgT601 GTGTGATTTT TTATCCGAAT
CodY box 6 mm
gCTAAgTATTTAAGTTA TACTTCAAAT ATAAGACCTG GTACTAATTC CTGCTAAAAG TGTTCGTTTTTGA ATGCGCTTCC681
CodY box 6 mm -35(!) SigA promote(?) -10(!) CodY box 6 mm
761 TtgAAtTgTG CAAACTGACG GAAaacTtTc AAAATaACAA TTGACAATCG CATGGCAACC ATATATATTA AAtAcTaaCA
CodY box 4 mm site 1 (!) DNase I foot-printing
841 tAAcATTTCT TGATATTAAt TTTTtTcAAA AaTGTGCGAC TAATCGAAAA AATAAAACCA TTTAACGAAG GAGATAATGA
rli60 (185 nt non-coding RNA, a putative riboswitch)
921 CTTATGAAAA CGACCAAATC AGTCATTACA ATTTTATTAC TCTAGAAGGA CTTTGAGCAC TGTAGAAATT TACAGTAGTT
CodY box 4 mm CodY box 6 mm
Stem-loop (DG -13.1 kcal/mole) poly-U tail (?)
1001 TGAGTCCTGT TTACGTTAAA TGGGATTCTA GCAAAGCATC CCATTGTTTT CATCATTGGG GTGCTTTTTA TTTAGCTAGA
CodY box 6 mm Rho-independent terminator(?)
1081 TTTCGAGTTT TCAAGCATCG AAAAGCCATT ATCAAGCGAG CAGATACTTA ATCATATAAA TTAATGCCAC GCTATTTAGt
site 2 (weak) DNase I foot-printing
1161 gaaTTCTaAA AATTCAGTGT CGGCAAACAA TTCTTAATTA GAAATGGGGT AAAGTCATAT GCGTAGTGAC AAAATAAAAA
CodY BOX 5 mm RBS(?) >>> ilvD
Fig 11. L.monocytogenes rli60 genomic region. Regions marked with (?) are un-confirmed assumptions. Important notes:
'CodY site1' have recently been proven to binds CodY [7], palindromes at 3'UTR of rli60, RNA polymerase binding site
(-10, -35).
References
1. Bowman J., Bittencourt C. and Ross T. (2008). Differential Gene Expression of Listeria
monocytogenes During High Hydrostatic Pressure Processing. Micribiology: 462-463.
2. Sesto N., Wuhrtzel O., Archambaud C., Sorek R. and Cossart P. (2013). The excludon: a new
concept in bacterial antisense RNA-mediated gene regulation. Nature review, microbiology: 75-77.
3. Cochrane J.C., Strobel S. A. (2008). Riboswitch Effectors as Protein Enzyme Cofactors. RNA vol
14: 993-995.
4. Mellin J. R. and Cossart P. (2012). The Non-Coding RNA World of the Bacterial Pathogen
Listeria monocytogenes. RNA Biology 9: 372-378.
5. Block K. F., Hammond M. C. and Breaker R. R. (2010). Evidence for Widespread Gene Control
Function by the ydaO Riboswitch Candidate. Journal Of Bacteriology vol. 192: 3983-3986.
6. Garst A. D., Edwards A. L. and Batey R. T. (2013). Riboswitches: Structures and Mechanisms.
Cold Spring Harbor Laboratory Press.
7. Lobel L., Nadejda S., Borovok I., Ruppin E. and Herskovits A. A. (2012). Integrative Genomic
Analysis Identifies Isoleucine and CodY as Regulators of Listeria monocytogenes Virulence. PLOS
genetics vol. 8, issue 9.
8. Hain T., Steinweg C., Kuenne C. T. and Chakroborty T. (2006). Whole-Genome Sequence of
Listeria welshimeri Reveals Common Steps in Genome Reduction with Listeria innocua as Compared
to Listeria monocytogenes. Journal of Bacteriology Vol.188 No.21: 7405-7407.](https://image.slidesharecdn.com/a18316e7-86e9-49b9-a490-677ac65927dc-161006113259/85/rli60-project-FINAL-13-320.jpg)
