This laboratory project report details the expression and purification of the Clocl_2077 protein from Clostridium clariflavum for the purposes of crystallization and structural determination. The Clocl_2077 protein is a putative fused anti-sigma factor and sigma factor protein that may be involved in cellulose degradation regulation. The project involved cloning the Clocl_2077 gene, expressing it in E. coli, and purifying the protein using nickel affinity and size exclusion chromatography. Additional experiments were conducted to express and purify the CBM3b domain of the Clocl_1053 protein to test its ability to bind carbohydrates. The successful purification of Clocl_2077 paved the way for future crystallization

![1
Abstract
Life on Earth depends on photosynthesis, which results in production of plant biomass having
cellulose as the major component. The carbon cycle is closed primarily as a result of the action of
cellulose-utilizing microorganisms [1]. Thus, microbial cellulose utilization is responsible for one
of the largest material flows in the biosphere and is of interest in relation to analysis of carbon
flux at both local and global scales. Cellulosic materials are particularly attractive in this context
because of their relatively low cost and plentiful supply. Aloof, breaking cellulose is no easy task.
The central technological impediment to more widespread utilization of this important resource
is the general absence of low-cost technology for overcoming the recalcitrance of cellulosic
biomass. A promising strategy to overcome this impediment involves the production of
cellulolytic enzymes, hydrolysis of biomass, and fermentation of resulting sugars to desired
products in a single process step via a cellulolytic microorganism or consortium [1, 2]. Generally,
the degradation processes can be achieved by a macromolecular complex named Cellulosome- A
machine comprising different enzymes which can digest the biomass efficiently [3]. This project
will deal with the cellulosome-producing anaerobic bacteria- Clostridium clariflavum working of
expression, purifying, crystallization and diffraction parts of the Clocl_2077 protein.
The main results of this work are the obtaining of a pure protein solution of Clocl_2077 and the
construction of experiments for future crystallizations, in order to reach a further understanding
helping the main goal above.
A side job of this project was also to over expresses and purifying the CBM3b protein (part of
Clocl_1053) protein of C. clariflavum and to construct a carbohydrate-binding assay to the
purifying protein in order to examine the protein ability to bind carbohydrates.
Introduction
Cellulolytic clostridia are prominently represented among bacterial species. These
organisms are able to solubilize lignocellulose, and their high rates of cellulose
utilization make them candidates for consolidated bioprocessing applications [4]. In
particular, anaerobic cellulolytic clostridia which grow at thermophilic temperatures
is aclariflavumlostridiumCto break down lignocellulose very efficiently.ableare
within the family Clostridiaceae isolated fromClostridiumIIIClustershaperod
dalthough retarde-motile-type positive, nonGram,c sludgethermophilic anaerobi
st because of its. This species is of intere], 54[flagella are presentedperitrichos
Clostridiumorganismmodel cellulolyticthe well studysimilarity to
environmental isolates to break downity ofand for the abilthermocellum
-omecellulosis aC. clostridiumned,oAs mentihemicellulose in addition to cellulose.
designed for efficientcomplexellulosomecheT.producing anaerobic bacterium
degradation of plant cell-wall polysaccharides in general and cellulose in particular. It
consist of a central 'scaffoldin' subunit that incorporates the various enzymes into the
complex, anchors the complex onto the cell surface of the bacterium and targets the
complex to the substrate (figure1). A cellulosome's major component is the CBM](https://image.slidesharecdn.com/99d0724b-071c-4f8b-a9cc-d5fd80d1d5b9-161006112344/85/Clocl_2077-crystallization-FINAL-2-320.jpg)
![2
(Carbohydrate Binding Module). CBM is a family of proteins that share a discrete
fold which enables them to bind different carbohydrates. So far, 64 CBM families
have been identified, based on amino acid sequence similarity [6]. These families
feature a great diversity in ligand specificity. there are characterized CBMs that
recognize crystalline cellulose, non-crystalline cellulose, chitin, xylan, starch and ex.
[7]. An example that will later go through a carbohydrate binding assay- as a 'side job'
of this study- is CBM3b1
(family-3b CBM) which characterized by a module of
approximately 150 amino acids organized in a β-sandwich fold and has the ability to
glycosyl hydrolasesofvarietyaare known to organizeriflavumclaC.bind cellulose.
and other catalytic subunits outside of the cell by means of the cellulosome. In this
bacterium there are few different examples of a cellulosome architecture structural
proteins such as typeI and typeII cohesin-dockerin interaction (fighre1), in addition to
a changeable number of the subunits comprising the different macromolecules. In
terms of the anchoring proteins, 4 different structures have been identified containing
SLH2
(S-layer homology). In addition, in contrast to others cellulosomal
4haveclariflavumC.,modules are not very commonCBM2ganisms wheremicroor
(!) of these domains that also associated with variable modules. This specific and
C.wideness of theorder to show theelaborated description is important in
can becellulosome components. As a result, a numerous possibilitiesclariflavum
taken in account, in order to seek for the perfect combination to manipulate and
accomplish the best efficient carbohydrate degradation complex.
Figure13
.Simplified model of a typical cellulosome, based on the C. thermocellum paradigm. This figure represent diverse
interactions inside the complex. C. clariflavum also have different and wide possibilities installing the varied relationships
between the different sub-units. Note that not all the interactions described in the 'Introduction' section are presented.
1
Historically, the division of the family into subgroups (a, b and c) was based on minor sequence differences combined with the
fact that the known scaffoldin-borne CBM3s (subgroup a) from four different clostridia could be differentiated by the existence
of a distinctive Trp-containing. Subfamilies b and c lacked this loop and were further differentiated by the lack of the standard
aromatic binding residues in subfamily c [8].
It.archaea, as well as amongbacteriacommonly found incell envelopeis a part of the(SLH)(surface layer)homologylayer-S
2
.glycoproteinsorproteinsconsists of a monomolecular layer composed of identical
3
This figure is given due to a lack of such relevant data of this work's bacteria in order to convey a graphic clue of the different
interaction described.](https://image.slidesharecdn.com/99d0724b-071c-4f8b-a9cc-d5fd80d1d5b9-161006112344/85/Clocl_2077-crystallization-FINAL-3-320.jpg)
![3
Former studies of other bacteria revealed the presence of putative genes for proteins,
homologous to the anti-σ factor RsgI protein of Bacillus subtilis [9]. Some of these
genes contain upstream to the rsgI an open reading frame (ORF) encoding for σ
factor, sigI, that are expected to be co-transcribed, as in B. subtilis [10]. Some of the
RsgI containing proteins are different in their modular structure from those of B.
subtilis RsgIs. These proteins have additional domains at the C-terminal that are
predicted to be located and act outside the cell membrane as CBMs. The proposed
regulatory mechanism of the cellulosomal genes, states that alternative σ factors are
activated in response to the polysaccharides in the extracellular surroundings (Fig. 2)
[9]. Bioinformatic analyses have shown a presence of such putative protein which
comprises anti-σ and σ factors in the same ORF also in C. clariflavum [11]. In
addition, an exclusive new sequence was discovered during examination the C.
clariflavum genome, indicating a new fused type of protein which comprises both
anti-σ and σ factors. The C-terminus putative module of these fused protein
designated Early set (E_set) and the complete protein is RsgI-like E_Set
(Clocl_2077). This protein contains an N-terminal sigI-like region (Region (Re). A)
fused to RsgI-like region with a trans-membrane domain (Re. B) that has a putative
sensing module (Re. C) which consist the E_set module at its C-terminus (fig. 2, ii).
The whole general function of Clocl_2077 is still unknown but can be predicted,
hence is the relevant of this study. The origin of the most basic hypothesis must lays
on a solid ground. That's why the initial work, carried out in this project, is to over-
express, purifying and figure the partial 'long' protein's structure of Clocl_2077, in
order to give a profound knowledge of the interactions between the subunits relating
to the cellulose utilization that will hopefully shed some light on the process. By
doing that I'm hoping to achieve some basis for further research of the cellulose
degradation by C. clariflavum. In addition, in this work I'm dealing with the CBM3b
protein part of Clocl_1053 in order to test its carbohydrates binding ability.](https://image.slidesharecdn.com/99d0724b-071c-4f8b-a9cc-d5fd80d1d5b9-161006112344/85/Clocl_2077-crystallization-FINAL-4-320.jpg)

![5
(Lysogeny Broth) + Kanamycin plates were used in order to identify the
transformants colonies.
Fig3. The predicted pET-28a(+) vector. Note that just the Clocl_2077 insert is present. Clocl_1053 was cloned at the same way
and place and using of the same vector.
***Transformated E. coli bacteria which adopted the vector containing Clocl_1053
were provided as a gift from Dr. Oren Yaniv The cloning was done using the primers:
forward (cut by NdeI) 5'-GCACATATGTCTGTTAAGCTCGGTATGTACAA-3', reverse (cut by XhoI)
5'-GCACTCGAGTTAGGGTTCAGTACCCCATAC-3'.
:2077 only)_(CloclssayAdetection by colony PCRectorV:STAGE2
4 colonies were chosen for colony PCR using the original primers described earlier. A
colony that received the vector will yield a ~735bp bend after electrophoresis in
Agarose gel, which refers to the Clocl_2077 gene's length (including the primers).
:1053 separately)_2077 & Clocl_locl(Cein expressionProt:STAGE3
4 different pre-cultures (starters) of E. coli harboring the expression plasmid were
cultivated. A gentle touch of a chosen colony (which showed a positive figure at the
colony-PCR assay) was mix with 5ml LB and 5µl kanamycin 50µg/ml for expression
at 310ºK, 270rpm rotation, O.N. Day afterwards each starter was grown under aerobic
conditions at 310ºK, 270rpm rotation 24h in TB (Terrific Broth) medium The
specifics growing conditions were chosen due to some results of previously studies [6,
7, 8].](https://image.slidesharecdn.com/99d0724b-071c-4f8b-a9cc-d5fd80d1d5b9-161006112344/85/Clocl_2077-crystallization-FINAL-6-320.jpg)



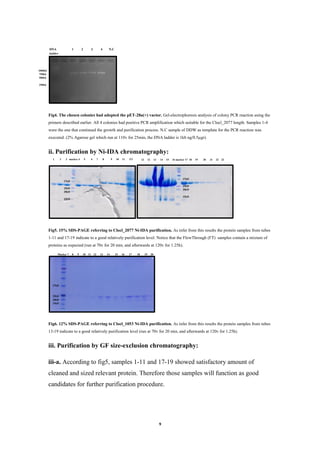


![12
Discussion
Based on the concentration value of Clocl_2077 reached in this project, I can assume
that the purification methods which had been chosen are suitable and the expression
and purification stages were executed satisfactory. As for this writing, though, no
crystals were observed. Therefore some explanation must be provided. First, the 'long'
protein of Clocl_2077 is an unknown module and perhaps not enough time for
crystallization may be the cause for getting no crystals. Second, and inevitably, there
is a good chance of different possible options existence for handling the whole
procedure which can be involves in successful crystallization. Therefore this section
will deal with possible improvements or alternatives of the different stages of
expression, purification and crystallization procedures.
The 'long protein' composes of two different modules: one partially trans-membrane
protein (anti-σ) and the other is outer cell protein (E-set). Therefore, when dealing
with membrane protein (MP) (or pseudo-membrane protein) analysis special acts
must be execute to achieve reliability result. E. coli is a popular host for over
expression due to, among others, its well understood genetics and rapid growth [12].
However, as with other expression systems, high-level MP production is typically
toxic to the cell and the yields of biologically active material are generally poor.
Based on the observation that the over expression of MPs in E. coli leads to their
aggregation and to reduce levels of host membrane and secretory proteins [12], it has
been suggested that special E. coli strain which will aid in properly MP expression is
needed. Previously studies shows that when expression of a pure membrane protein
was induced in BL21(DE3) E. coli strain (just as used in this project), most of the
BL21(DE3) host cells died. Similar effects were also observed with expression
vectors for 10 globular proteins (GP). Therefore, protein over-production in this
expression system is either limited or prevented by bacterial cell death. Out of the few
survivors of BL21(DE3) a mutant host C41(DE3) was selected that grew to high
saturation cell density, and produced the protein as inclusion bodies at an elevated
level without toxic effect. Some proteins that were expressed poorly in BL21(DE3),
and others where the toxicity of the expression plasmids prevented transformation
into this host, were also over-produced successfully in C41(DE3). The examples
include GPs as well as MPs, and therefore, strain C41(DE3) is generally superior to
BL21(DE3) as a host for MP over-expression [13]. The final concentration of the](https://image.slidesharecdn.com/99d0724b-071c-4f8b-a9cc-d5fd80d1d5b9-161006112344/85/Clocl_2077-crystallization-FINAL-13-320.jpg)
![13
protein used for crystallization was relatively poor (in spite of what was written at the
beginning of this section) and this special strain perhaps may grant a better one.
In addition, the pET E. coli expression vector that was used in this project which is a
T7 RNA polymerase promoter driven and IsoPropyl-b-D-ThioGalactopyranoside
(IPTG) inducible are useful tool for the generation of expression constructs.
Alternatively, the pBAD vector system for E. coli expression which uses arabinose
induction has been implemented successfully for the production of MPs for X-ray
studies. Supporting studies to this assumption had found tight regulation, modulation,
and high-level expression of MPs by vectors containing the Arabinose PBAD
Promoter [14, 15].
Another issue to consider concerning the dis-crystallization is the vital isolation of
membrane fraction from MP during protein preparation. Diffraction quality crystals
are particularly difficult to prepare currently when a membrane source is used. The
reason for this is our limited ability to manipulate proteins bearing
hydrophobic/amphiphilic surfaces that are usually enveloped with membrane lipid.
More often than not, the protein gets trapped as an intractable aggregate in its watery
course from membrane to crystal. As a result, access to the structure, and thus
function is limited. Hence, for purification and crystallization, MPs need to be
extracted from the lipid membrane in which they were expressed using a special
detergent. For most expression systems, this extraction is performed on the isolated
membrane fraction but can be extracted from whole cells [16]. Whether solubilizing
from membranes or from whole cells, the goal is similar- to yield a water-soluble
Protein–Detergent–Lipid Complex (PDLC) (Fig11),Which will further lose the lipids
component and yield Protein-Detergent Complex (PDC). The identification and
desirable concentration of the detergent most suitable for a particular protein target is
an empirical process, when the ideal detergent extract all of the membrane protein
target from the membrane, maintains the native fold of the protein and forms a PDC
that is stable throughout purification and crystallization [16].](https://image.slidesharecdn.com/99d0724b-071c-4f8b-a9cc-d5fd80d1d5b9-161006112344/85/Clocl_2077-crystallization-FINAL-14-320.jpg)


