Poster_GCP_Knapp
•
0 likes•53 views
1) Researchers developed a pipeline to build and annotate a Sanger-454 transcript assembly database for groundnut, containing over 100,000 unigenes. 2) They mined this EST database to identify over 7,000 perfect simple sequence repeats (SSRs), selecting around 2,100 to develop into EST-SSR markers. 3) A pilot study screening 58 EST-SSR markers on elite cultivars and wild relatives found around half were polymorphic among cultivars but diversity was low, while 80% were polymorphic between wild relatives, demonstrating their greater utility for genetic mapping.
Report
Share
Report
Share
Download to read offline
Recommended
Comparative differential leucocyte count and morphometrical analyses of black...

Comparative differential leucocyte count and morphometrical analyses of black...African Journal of Biological Sciences
This paper aims at a systematic approach to morphologically characterize of five types of white blood cells (WBC), and its nuclei from light microscopic image of blood samples. Hence, cellular and nuclei based geometric features are computed and analyzed statistically with t-test to show their discriminating potentiality among the species. In morphometry study, the length and breadth along with nucleus of leukocytes are compared between and within the species using oneway Analysis of Variance (ANOVA) followed by Tukey’s pairwise comparison tests. In this study, the estimated values of Rattus rattus and Rattus norvegicus with respect to sex were compared. A total of 20 black and white rats (05 each from males and females) were collected. Blood samples were then collected from the caudal vein of anaesthetized rats. In differential leucocyte count, the parameters namely, lymphocyte, monocyte, neutrophil (p < 0.001) and eosinophil and basophil (p < 0.05) reveal significant difference. In morphometrical study, the cell length, breadth along with nucleus of lymphocyte, monocyte, neutrophil (p < 0.01) and eosinophil, basophil (p < 0.05) deviates significantly between and within the species.Hematological Parameters of three Strains of Local Cocks in Northern Nigeria

The study was conducted to determine the hematological parameters of three strains of the Nigerian indigenous cocks. A total of 15 sexually matured (14-18 month of age) breeders cocks comprising (5 normal feathered, 5 frizzled feathered and 5 naked neck) were used for the experiment. The study was conducted from October to December 2016 at the Teaching and Research Farm University of Maiduguri. Blood samples were collected from 9 breeder’s cocks which were randomly selected 3 per genotype and used for hematological parameters examination. Hematological examination such as Packed Cell Volume (PCV), Red Blood Cell ( RBC) , Haemoglobin (Hb), White Blood Cell (WBC), Mean Corpuscular Haemoglobin concentration ( MCHC), Mean Corpuscular Haemoglobin (MCH) and Mean Corpuscular Volume ( MCV) showed significant (P<0.05)> 0.05) different between normal feathered and frizzle feathered but there is significant difference ( P< 0.05) with naked necked cock. Neutrophil ( N) showed significant (P<0.05)>0.05) difference between normal feathered and naked neck feathered , fizzle feathered and necked neck respectively but showed significant (P<0.05) difference between fizzle feathered and normal feathered respectively for M and E. the study concluded that variation in the heamatoloical parameters between three strains of local chicken in Nigeria is due to difference in their genetic makeup.
D03011027030

The IOSR Journal of Pharmacy (IOSRPHR) is an open access online & offline peer reviewed international journal, which publishes innovative research papers, reviews, mini-reviews, short communications and notes dealing with Pharmaceutical Sciences( Pharmaceutical Technology, Pharmaceutics, Biopharmaceutics, Pharmacokinetics, Pharmaceutical/Medicinal Chemistry, Computational Chemistry and Molecular Drug Design, Pharmacognosy & Phytochemistry, Pharmacology, Pharmaceutical Analysis, Pharmacy Practice, Clinical and Hospital Pharmacy, Cell Biology, Genomics and Proteomics, Pharmacogenomics, Bioinformatics and Biotechnology of Pharmaceutical Interest........more details on Aim & Scope).
All manuscripts are subject to rapid peer review. Those of high quality (not previously published and not under consideration for publication in another journal) will be published without delay.
Assessment of Genetic Diversity in 13 Local Banana (Musa Spp.) Cultivars Usin...

Abstract: A Study was conducted to investigate the genetic variability among 13 local banana cultivars using 3 SSR primers of Mb1-69, Mb1-113 and Mb1-134. All the primer pairs amplified a total of 29 different marker bands with an average of 9.6 bands per primer. Among the 29 bands only 4 bands were monomorphic and the rest 25 bands were polymorphic. The sizes of the amplified DNA bands in 13 local banana cultivars varied from 200 bp to 600 bp. The primer Mb1-113 amplified the highest (14) number of DNA bands and the primer Mb1-69 amplified the lowest (7) number of DNA bands whilst primer Mb1-134 amplified 8 DNA bands. The values of pair-wise genetic distances ranged from 1.00 to 9.00 indicating the presence of wide genetic diversity. The dendogram constructed based on phylogenetic relationship analysis revealed that the highest genetic diversity (9.00) found between the cultivars champa and jawayta and also the cultivars champa and jahazy whilst the lowest (1.00) between the cultivars doubled haploid and kathaly, doubled haploid and sorishafruity, doubled haploid and amritsagor and doubled haploid and ganasundory. The UPGMA dendogram has segregated the 13 local banana cultivars into two major clusters. Agnishwar and champa formed in cluster 1 and the rest of the cultivars like sobri jesore, sobri, anazy, kathaly, jawayta, sorishafruity, amritsagor, jahazy, bangle, ganasundory and doubled haploid have constituted the cluster 2.
Major biological nucleotide databases

GenBank (Genetic Sequence Databank),European Molecular Biology Laboratory (EMBL)
Analysis of some Capparis L. accessions from Turkey based on IRAP and seed pr...

Analysis of some Capparis L. accessions from Turkey based on IRAP and seed pr...Agriculture Journal IJOEAR
Abstract— 15 accessions from 10 different grid square of Turkey were analysed based on IRAP and seed protein patterns in order to observe the genetic diversity in the gene pool of Capparis. High levels of polymorphisms were detected with IRAP primers (93%) and seed protein electrophoresis (55.5%). Specific delineation between C. spinosa and C. ovata, and segregations of the accessions related to infraspecific status and eco-geographical distributions were presented in the dendrograms and PCA analysis. Significantly correlation between IRAP markers and seed protein profiles of the specimens was detected (p< 0.0001). Combination of genomic/proteomic marker systems may be useful approach for determining the broad genetic diversity in gene pool of Capparis, identification of the germplasms and ecologically tolerant genotypes in breeding programs.Recommended
Comparative differential leucocyte count and morphometrical analyses of black...

Comparative differential leucocyte count and morphometrical analyses of black...African Journal of Biological Sciences
This paper aims at a systematic approach to morphologically characterize of five types of white blood cells (WBC), and its nuclei from light microscopic image of blood samples. Hence, cellular and nuclei based geometric features are computed and analyzed statistically with t-test to show their discriminating potentiality among the species. In morphometry study, the length and breadth along with nucleus of leukocytes are compared between and within the species using oneway Analysis of Variance (ANOVA) followed by Tukey’s pairwise comparison tests. In this study, the estimated values of Rattus rattus and Rattus norvegicus with respect to sex were compared. A total of 20 black and white rats (05 each from males and females) were collected. Blood samples were then collected from the caudal vein of anaesthetized rats. In differential leucocyte count, the parameters namely, lymphocyte, monocyte, neutrophil (p < 0.001) and eosinophil and basophil (p < 0.05) reveal significant difference. In morphometrical study, the cell length, breadth along with nucleus of lymphocyte, monocyte, neutrophil (p < 0.01) and eosinophil, basophil (p < 0.05) deviates significantly between and within the species.Hematological Parameters of three Strains of Local Cocks in Northern Nigeria

The study was conducted to determine the hematological parameters of three strains of the Nigerian indigenous cocks. A total of 15 sexually matured (14-18 month of age) breeders cocks comprising (5 normal feathered, 5 frizzled feathered and 5 naked neck) were used for the experiment. The study was conducted from October to December 2016 at the Teaching and Research Farm University of Maiduguri. Blood samples were collected from 9 breeder’s cocks which were randomly selected 3 per genotype and used for hematological parameters examination. Hematological examination such as Packed Cell Volume (PCV), Red Blood Cell ( RBC) , Haemoglobin (Hb), White Blood Cell (WBC), Mean Corpuscular Haemoglobin concentration ( MCHC), Mean Corpuscular Haemoglobin (MCH) and Mean Corpuscular Volume ( MCV) showed significant (P<0.05)> 0.05) different between normal feathered and frizzle feathered but there is significant difference ( P< 0.05) with naked necked cock. Neutrophil ( N) showed significant (P<0.05)>0.05) difference between normal feathered and naked neck feathered , fizzle feathered and necked neck respectively but showed significant (P<0.05) difference between fizzle feathered and normal feathered respectively for M and E. the study concluded that variation in the heamatoloical parameters between three strains of local chicken in Nigeria is due to difference in their genetic makeup.
D03011027030

The IOSR Journal of Pharmacy (IOSRPHR) is an open access online & offline peer reviewed international journal, which publishes innovative research papers, reviews, mini-reviews, short communications and notes dealing with Pharmaceutical Sciences( Pharmaceutical Technology, Pharmaceutics, Biopharmaceutics, Pharmacokinetics, Pharmaceutical/Medicinal Chemistry, Computational Chemistry and Molecular Drug Design, Pharmacognosy & Phytochemistry, Pharmacology, Pharmaceutical Analysis, Pharmacy Practice, Clinical and Hospital Pharmacy, Cell Biology, Genomics and Proteomics, Pharmacogenomics, Bioinformatics and Biotechnology of Pharmaceutical Interest........more details on Aim & Scope).
All manuscripts are subject to rapid peer review. Those of high quality (not previously published and not under consideration for publication in another journal) will be published without delay.
Assessment of Genetic Diversity in 13 Local Banana (Musa Spp.) Cultivars Usin...

Abstract: A Study was conducted to investigate the genetic variability among 13 local banana cultivars using 3 SSR primers of Mb1-69, Mb1-113 and Mb1-134. All the primer pairs amplified a total of 29 different marker bands with an average of 9.6 bands per primer. Among the 29 bands only 4 bands were monomorphic and the rest 25 bands were polymorphic. The sizes of the amplified DNA bands in 13 local banana cultivars varied from 200 bp to 600 bp. The primer Mb1-113 amplified the highest (14) number of DNA bands and the primer Mb1-69 amplified the lowest (7) number of DNA bands whilst primer Mb1-134 amplified 8 DNA bands. The values of pair-wise genetic distances ranged from 1.00 to 9.00 indicating the presence of wide genetic diversity. The dendogram constructed based on phylogenetic relationship analysis revealed that the highest genetic diversity (9.00) found between the cultivars champa and jawayta and also the cultivars champa and jahazy whilst the lowest (1.00) between the cultivars doubled haploid and kathaly, doubled haploid and sorishafruity, doubled haploid and amritsagor and doubled haploid and ganasundory. The UPGMA dendogram has segregated the 13 local banana cultivars into two major clusters. Agnishwar and champa formed in cluster 1 and the rest of the cultivars like sobri jesore, sobri, anazy, kathaly, jawayta, sorishafruity, amritsagor, jahazy, bangle, ganasundory and doubled haploid have constituted the cluster 2.
Major biological nucleotide databases

GenBank (Genetic Sequence Databank),European Molecular Biology Laboratory (EMBL)
Analysis of some Capparis L. accessions from Turkey based on IRAP and seed pr...

Analysis of some Capparis L. accessions from Turkey based on IRAP and seed pr...Agriculture Journal IJOEAR
Abstract— 15 accessions from 10 different grid square of Turkey were analysed based on IRAP and seed protein patterns in order to observe the genetic diversity in the gene pool of Capparis. High levels of polymorphisms were detected with IRAP primers (93%) and seed protein electrophoresis (55.5%). Specific delineation between C. spinosa and C. ovata, and segregations of the accessions related to infraspecific status and eco-geographical distributions were presented in the dendrograms and PCA analysis. Significantly correlation between IRAP markers and seed protein profiles of the specimens was detected (p< 0.0001). Combination of genomic/proteomic marker systems may be useful approach for determining the broad genetic diversity in gene pool of Capparis, identification of the germplasms and ecologically tolerant genotypes in breeding programs.Assesment of genetic divergence in chickpea kabuli cultivars

ASSESMENT OF GENETIC DIVERGENCE IN CHICKPEA KABULI CULTIVARS
Biological database by kk sahu

INTRODUCTION
HISTORY
WHAT ARE THE DATABASE…?
WHY DATABASE….?
THE “PERFECT” DATABASE
IDENTIFIERS and ACCESSION NUMBER
TECHNICAL DESIGN
MAINTAINANCE OF BIOLOGICAL DATABASES..
GENERAL FEATURES
SOURCES OF BIOLOGICAL DATA…
DIFFERENT TYPES OF BIOLOGICAL DATABASE
FUNCTION
DATA ENTRY AND QUALITY CONTROL
AVAILIBILITY
APPLICATION
DATA RECORD AT THE YEAR 2004
CONCLUSION
REFFERENCES
PAG Poster

Poster presented at Plant and Animal Genome Conference 2007. Concerns development and deployment of EST-SSRs in peanut.
Yam: Recent Advances in Genetic and Genomic Resources 

Yam: Recent Advances in Genetic and Genomic Resources CGIAR Research Program on Roots, Tubers and Bananas
Presented at the CGIAR Research Program on Roots, Tubers and Bananas (RTB) Annual Meeting from 8-10 December 2015 in Lima, Peru.Gene expression group presentation at GAW 19

Presentation slides of the gene expression group at GAW19, Genetic Analysis Workshop, Vienna 2014
Building Genomic Data Processing and Machine Learning Workflows Using Apache ...

Epinomics is advancing epigenetic research to drive personalized medicine, using epigenomic data analysis. Their goal is to provide an analysis resource to the community that will promote high-quality data and replicable and interpretable results. They work with academic and commercial users to ingest and analyze their genomic sequencing data and metadata. They extract epigenetic features from the sequenced genome, called “chromatin accessibility”, which are indicative of instrumental epigenetic changes responsible for differential gene expression and disease development.
Epinomics has built an Apache Spark-based pipeline that retrieves chromatin accessibility data from the epigenome, uses GraphX to find overlapping accessibility atlas and then clusters the data and runs machine learning algorithms. This session will provide a primer on epigenomics, details about Epinomics’ Spark-based data pipeline focusing on parallel bioinformatic analysis, and how they use machine learning models to build the epigenomic landscape and accelerate the field of personalized immunotherapy. use GraphX to find overlapping accessibility atlas and then cluster the data and run machine learning algorithms.
In this talk we will provide a primer on epigenomics, details about our Spark based data pipeline focusing on parallel bioinformatic analysis and how we use machine learning models to build the epigenomic landscape and accelerate the field of personalized immunotherapy.
Systemic analysis of data combined from genetic qtl's and gene expression dat...

Systemic analysis of data combined from genetic qtl's and gene expression dat...Laurence Dawkins-Hall
Elucidating changes in gene expression by Micro array genomic sweeps of genetic QTLs linked to Tryp resistance in WT cattle to identify putative candidates underpinning pathophysiology 21 kebere bezaweletaw 207-217

The International Institute for Science, Technology and Education (IISTE) , International Journals Call for papaers: http://www.iiste.org/Journals
A systematic, data driven approach to the combined analysis of microarray and...

A systematic, data driven approach to the combined analysis of microarray and...Laurence Dawkins-Hall
The use of gene expression data from Micro arrays coupled with WT QTL's linked to Tryp resistance phenotypes in Cattle to elucidate pertinent genetic changes underpinning phenotype in putative candidate genes Data as research output, data as part of the scholarly record

Talk given at SciELO15, 24 October 2013, São Paulo Brazil. Video followed by slides.
Frequency of Polyploids of Solanum tuberosum Dihaploids in 2X × 2X Crosses

Frequency of Polyploids of Solanum tuberosum Dihaploids in 2X × 2X CrossesJournal of Agriculture and Crops
When breeding diploid potatoes, tetraploid progeny can result from the union of 2n eggs and 2n pollen in 2x-2x crosses. Thirty-three crosses were made to examine tetraploid progeny frequency in 2x-2x crosses. All crosses were between S. tuberosum dihaploids and diploid self-compatible donors, M6 and DRH S6-10-4P17. Using chloroplast counting for ploidy determination, the frequency of tetraploid progeny was as high as 45% in one of the 33 crosses. Based upon single nucleotide polymorphism (SNP) genotyping, the tetraploid progeny were attributed to bilateral sexual polyploidization (BSP), which is caused by the union of 2n egg and 2n pollen. Dihaploids were identified that produce lower frequencies of 2n eggs. The results of this study suggest that S. tuberosum dihaploids with a high frequency of 2n eggs should be avoided in 2x - 2x crosses for diploid breeding programs.More Related Content
What's hot
Assesment of genetic divergence in chickpea kabuli cultivars

ASSESMENT OF GENETIC DIVERGENCE IN CHICKPEA KABULI CULTIVARS
Biological database by kk sahu

INTRODUCTION
HISTORY
WHAT ARE THE DATABASE…?
WHY DATABASE….?
THE “PERFECT” DATABASE
IDENTIFIERS and ACCESSION NUMBER
TECHNICAL DESIGN
MAINTAINANCE OF BIOLOGICAL DATABASES..
GENERAL FEATURES
SOURCES OF BIOLOGICAL DATA…
DIFFERENT TYPES OF BIOLOGICAL DATABASE
FUNCTION
DATA ENTRY AND QUALITY CONTROL
AVAILIBILITY
APPLICATION
DATA RECORD AT THE YEAR 2004
CONCLUSION
REFFERENCES
What's hot (10)
Assesment of genetic divergence in chickpea kabuli cultivars

Assesment of genetic divergence in chickpea kabuli cultivars
Similar to Poster_GCP_Knapp
PAG Poster

Poster presented at Plant and Animal Genome Conference 2007. Concerns development and deployment of EST-SSRs in peanut.
Yam: Recent Advances in Genetic and Genomic Resources 

Yam: Recent Advances in Genetic and Genomic Resources CGIAR Research Program on Roots, Tubers and Bananas
Presented at the CGIAR Research Program on Roots, Tubers and Bananas (RTB) Annual Meeting from 8-10 December 2015 in Lima, Peru.Gene expression group presentation at GAW 19

Presentation slides of the gene expression group at GAW19, Genetic Analysis Workshop, Vienna 2014
Building Genomic Data Processing and Machine Learning Workflows Using Apache ...

Epinomics is advancing epigenetic research to drive personalized medicine, using epigenomic data analysis. Their goal is to provide an analysis resource to the community that will promote high-quality data and replicable and interpretable results. They work with academic and commercial users to ingest and analyze their genomic sequencing data and metadata. They extract epigenetic features from the sequenced genome, called “chromatin accessibility”, which are indicative of instrumental epigenetic changes responsible for differential gene expression and disease development.
Epinomics has built an Apache Spark-based pipeline that retrieves chromatin accessibility data from the epigenome, uses GraphX to find overlapping accessibility atlas and then clusters the data and runs machine learning algorithms. This session will provide a primer on epigenomics, details about Epinomics’ Spark-based data pipeline focusing on parallel bioinformatic analysis, and how they use machine learning models to build the epigenomic landscape and accelerate the field of personalized immunotherapy. use GraphX to find overlapping accessibility atlas and then cluster the data and run machine learning algorithms.
In this talk we will provide a primer on epigenomics, details about our Spark based data pipeline focusing on parallel bioinformatic analysis and how we use machine learning models to build the epigenomic landscape and accelerate the field of personalized immunotherapy.
Systemic analysis of data combined from genetic qtl's and gene expression dat...

Systemic analysis of data combined from genetic qtl's and gene expression dat...Laurence Dawkins-Hall
Elucidating changes in gene expression by Micro array genomic sweeps of genetic QTLs linked to Tryp resistance in WT cattle to identify putative candidates underpinning pathophysiology 21 kebere bezaweletaw 207-217

The International Institute for Science, Technology and Education (IISTE) , International Journals Call for papaers: http://www.iiste.org/Journals
A systematic, data driven approach to the combined analysis of microarray and...

A systematic, data driven approach to the combined analysis of microarray and...Laurence Dawkins-Hall
The use of gene expression data from Micro arrays coupled with WT QTL's linked to Tryp resistance phenotypes in Cattle to elucidate pertinent genetic changes underpinning phenotype in putative candidate genes Data as research output, data as part of the scholarly record

Talk given at SciELO15, 24 October 2013, São Paulo Brazil. Video followed by slides.
Frequency of Polyploids of Solanum tuberosum Dihaploids in 2X × 2X Crosses

Frequency of Polyploids of Solanum tuberosum Dihaploids in 2X × 2X CrossesJournal of Agriculture and Crops
When breeding diploid potatoes, tetraploid progeny can result from the union of 2n eggs and 2n pollen in 2x-2x crosses. Thirty-three crosses were made to examine tetraploid progeny frequency in 2x-2x crosses. All crosses were between S. tuberosum dihaploids and diploid self-compatible donors, M6 and DRH S6-10-4P17. Using chloroplast counting for ploidy determination, the frequency of tetraploid progeny was as high as 45% in one of the 33 crosses. Based upon single nucleotide polymorphism (SNP) genotyping, the tetraploid progeny were attributed to bilateral sexual polyploidization (BSP), which is caused by the union of 2n egg and 2n pollen. Dihaploids were identified that produce lower frequencies of 2n eggs. The results of this study suggest that S. tuberosum dihaploids with a high frequency of 2n eggs should be avoided in 2x - 2x crosses for diploid breeding programs.Genotyping an invasive vine

The climbing vine kudzu, a member of the leguminous
pea family (Fabaceae), was introduced into the USA
from its native Asia in the 1800s. It was initially lauded
for efficacy in erosion control along highways and as a
high-quality grazing crop for livestock. P. montana var.
lobata has since become a truculent invasive, spreading
via vegetative runners and seed dispersal. Seven
million acres of the American southeast are now
plagued by this vine.
Genome responses of trypanosome infected cattle

Genomic gene expression changes resulting from Trypanosomiasis: a horizontal study Examining expression changes elucidated by micro arrays in seminal tissues associated with the pathophysiology of Trypanosomiasis during disease progression
Whole Transcriptome Analysis of Testicular Germ Cell Tumors

Next generation sequencing of the whole transcriptome enables high resolution measurement of gene expression activity in different tissue and cell types. This methodology provides an in depth study of known transcripts and depending on the data analysis, allows identification of additional transcript types such as transcript variants, fusion transcripts, and small and long ncRNAs.
In this study we performed RNA-Seq using the Ion Torrent™ sequencing platform to compare the expression profile of testicular germ cell cancers (seminoma type, n=3) and normal testis (n=3). Using Partek Flow® 3.0 and TopHat/BowTie or Star aligners, we aligned the reads to the human genome and mapped sequences to the RefSeq database. Differentially expressed genes were identified and screened with additional germ cell tumors.
PCA analysis showed clear separation of the two sample types indicating biological differences. List of differentially expressed genes generated from TopHat/Bowtie and Star were similar. We identified a large number of genes that were up and down regulated with high degree of significance (p<0.01,>2X FC (fold change)). These included genes related to testicular tissue type, stem cell pluripotency (NANOG; POU5F1) and proliferation (KRAS, CCND2).
In addition, a number of differentially expressed noncoding RNAs were identified (SNORD12B, XIST). The method was validated on a small set of genes (n=20) using qPCR (TaqMan® Assays) and were found to be correlated. We used the OpenArray® platform to quickly and quantitatively screen 102 differentially expressed genes and 10 endogenous control genes across a number of different testicular germ cell cancer types.
We used a complete work flow solution from sample prep to NGS to qPCR to compare the expression profile of normal testis and seminoma type germ cell tumors. From the NGS experiments we identified a large number of differentially expressed genes for qPCR screening with samples from different types of germ cell tumors. Results from these screening studies will be presented.
Molecular phylogenetics

This presentation entitled 'Molecular phylogenetics and its application' deals with all the developmental ideas and basics in the field of bioinformatics.
Striving for excellence in yam breeding using genomics tools

Striving for excellence in yam breeding using genomics toolsInternational Institute of Tropical Agriculture
Leveraging Genomic tools and pilelines to achieve reasonable progress in yam breeding and genomics.Similar to Poster_GCP_Knapp (20)
Yam: Recent Advances in Genetic and Genomic Resources 

Yam: Recent Advances in Genetic and Genomic Resources
Genetic diversity in wild and cultivated peanut_Khanal_2008

Genetic diversity in wild and cultivated peanut_Khanal_2008
Building Genomic Data Processing and Machine Learning Workflows Using Apache ...

Building Genomic Data Processing and Machine Learning Workflows Using Apache ...
Systemic analysis of data combined from genetic qtl's and gene expression dat...

Systemic analysis of data combined from genetic qtl's and gene expression dat...
A systematic, data driven approach to the combined analysis of microarray and...

A systematic, data driven approach to the combined analysis of microarray and...
Data as research output, data as part of the scholarly record

Data as research output, data as part of the scholarly record
Frequency of Polyploids of Solanum tuberosum Dihaploids in 2X × 2X Crosses

Frequency of Polyploids of Solanum tuberosum Dihaploids in 2X × 2X Crosses
Whole Transcriptome Analysis of Testicular Germ Cell Tumors

Whole Transcriptome Analysis of Testicular Germ Cell Tumors
Striving for excellence in yam breeding using genomics tools

Striving for excellence in yam breeding using genomics tools
More from Sameer Khanal
Linkage mapping lab

This update provides functioning links for downloading mapmaker. Please note that the website provides python codes for automating some of the most manual functions used in mapmaker.
Transcriptomics of RKN resitance in Upland Cotton

Presented at Institute of Plant Breeding, Genetics and Genomics (IPBGG) retreat in May 2019.
Effects of exotic alleles and genetic backgrounds on fiber quality traits in ...

Jeevan Adhikari's PAG XXVI poster
Dissecting quantitative variation introgressed into bermudagrass and Upland c...

Sameer Khanal's dissertation defense slides (April 23, 2018). Results redacted!
Microsatellite markers in bermudagrass

Used TetraploidMap to build linkage groups, results were not complete. Used other approaches instead.
Cytogenetics of arachis_spp.

This work was done by Sameer Khanal as a part of project requirement for course The work was done by Sameer Khanal as a project requirement of course "Plant Cytogenetics: Behavior and Evolution of the Plant Genome" [PBGG(CRSS) 8890] at the University of Georgia.
Linkage mapping and QTL analysis_Lab

Undergraduate level introductory laboratory exercise on linkage mapping and QTL analysis in experimental populations. Prepared in 2015.
Linkage mapping and QTL analysis_Lecture

Undergraduate level introductory lecture on linkage mapping and QTL analysis in experimental populations. Prepared in 2015.
Winning the future with GMOs

Prepared as a part of assignments for PBGG seminar @ UGA. Discusses misinformation regarding GMOs, backs up assertions with scientific evidences. Target audience: general public. Prepared and presented in 2013.
Sunflower domestication khanal_2010

A seminar presentation as a part of PBIO seminar at UGA discussing diverse school of thoughts about sunflower domestication. Prepared and presented in 2010.
Comfy cotton clothes and their genetics khanal 2013

A presentation on cotton genetics and breeding tailored to general audience as a part of a communication seminar at UGA.
More from Sameer Khanal (20)
Effects of exotic alleles and genetic backgrounds on fiber quality traits in ...

Effects of exotic alleles and genetic backgrounds on fiber quality traits in ...
Dissecting quantitative variation introgressed into bermudagrass and Upland c...

Dissecting quantitative variation introgressed into bermudagrass and Upland c...
Comfy cotton clothes and their genetics khanal 2013

Comfy cotton clothes and their genetics khanal 2013
Poster_GCP_Knapp
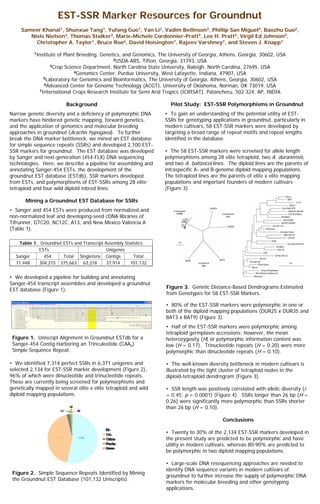
- 1. • We developed a pipeline for building and annotating Sanger-454 transcript assemblies and developed a groundnut EST database (Figure 1). EST-SSR Marker Resources for Groundnut Sameer Khanal1, Shunxue Tang1, Yufang Guo1, Yan Li2, Vadim Beilinson3, Phillip San Miguel4, Baozhu Guo2, Niels Nielsen3, Thomas Stalker3, Marie-Michele Cordonnier-Pratt5, Lee H. Pratt5, Virgil Ed Johnson5, Christopher A. Taylor1, Bruce Roe6, David Hoisington7, Rajeev Varshney7, and Steven J. Knapp1 1Institute of Plant Breeding, Genetics, and Genomics, The University of Georgia, Athens, Georgia, 30602, USA 2USDA-ARS, Tifton, Georgia, 31793, USA 3Crop Science Department, North Carolina State University, Raleigh, North Carolina, 27695, USA 4Genomics Center, Purdue University, West Lafayette, Indiana, 47907, USA 5Laboratory for Genomics and Bioinformatics, The University of Georgia, Athens, Georgia, 30602, USA 6Advanced Center for Genome Technology (ACGT), University of Oklahoma, Norman, OK 73019, USA 7International Crops Research Institute for Semi Arid Tropics (ICRISAT), Patancheru, 502 324, AP, INDIA Background Narrow genetic diversity and a deficiency of polymorphic DNA markers have hindered genetic mapping, forward genetics, and the application of genomics and molecular breeding approaches in groundnut (Arachis hypogaea). To further break the DNA marker bottleneck, we mined an EST database for simple sequence repeats (SSRs) and developed 2,100 EST- SSR markers for groundnut. The EST database was developed by Sanger and next-generation (454-FLX) DNA sequencing technologies. Here, we describe a pipeline for assembling and annotating Sanger-454 ESTs, the development of the groundnut EST database (ESTdb), SSR markers developed from ESTs, and polymorphisms of EST-SSRs among 28 elite tetraploid and four wild diploid inbred lines. Mining a Groundnut EST Database for SSRs • Sanger and 454 ESTs were produced from normalized and non-normalized leaf and developing-seed cDNA libraries of Tifrunner, GTC20, NC12C, A13, and New Mexico Valencia A (Table 1). Table 1. Groundnut ESTs and Transcript Assembly Statistics ESTs Unigenes Sanger 454 Total Singletons Contigs Total 71,448 304,215 375,663 63,218 37,914 101,132 Figure 1. Uniscript Alignment in Groundnut ESTdb for a Sanger-454 Contig Harboring an Trinculeotide (CAA5) Simple Sequence Repeat. Figure 2. Simple Sequence Repeats Identified by Mining the Groundnut EST Database (101,132 Uniscripts). Pilot Study: EST-SSR Polymorphisms in Groundnut • To gain an understanding of the potential utility of EST- SSRs for genotyping applications in groundnut, particularly in modern cultivars, 58 EST-SSR markers were developed by targeting a broad range of repeat motifs and repeat lengths identified in the database. • The 58 EST-SSR markers were screened for allele length polymorphisms among 28 elite tetraploid, two A. duranensis, and two A. batizocoi lines. The diploid lines are the parents of intraspecific A- and B-genome diploid mapping populations. The tetraploid lines are the parents of elite x elite mapping populations and important founders of modern cultivars (Figure 3). • We identified 7,314 perfect SSRs in 6,371 unigenes and selected 2,134 for EST-SSR marker development (Figure 2), 96% of which were dinucleotide and trinucleotide repeats. These are currently being screened for polymorphisms and genetically mapped in several elite x elite tetraploid and wild diploid mapping populations. Figure 3. Genetic Distance-Based Dendrograms Estimated from Genotypes for 58 EST-SSR Markers. • 80% of the EST-SSR markers were polymorphic in one or both of the diploid mapping populations (DUR25 x DUR35 and BAT3 x BAT9) (Figure 3). • Half of the EST-SSR markers were polymorphic among tetraploid germplasm accessions; however, the mean heterozygosity (H) or polymorphic information content was low (H = 0.17). Trinucleotide repeats (H = 0.20) were more polymorphic than dinucleotide repeats (H = 0.10). • The well-known diversity bottleneck in modern cultivars is illustrated by the tight cluster of tetraploid nodes in the diploid-tetraploid dendrogram (Figure 3). • SSR length was positively correlated with allelic diversity (r = 0.45; p < 0.0001) (Figure 4). SSRs longer than 26 bp (H = 0.26) were significantly more polymorphic than SSRs shorter than 26 bp (H = 0.10). Conclusions • Twenty to 30% of the 2,134 EST-SSR markers developed in the present study are predicted to be polymorphic and have utility in modern cultivars, whereas 80-90% are predicted to be polymorphic in two diploid mapping populations. • Large-scale DNA resequencing approaches are needed to identify DNA sequence variants in modern cultivars of groundnut to further increase the supply of polymorphic DNA markers for molecular breeding and other genotyping applications.
