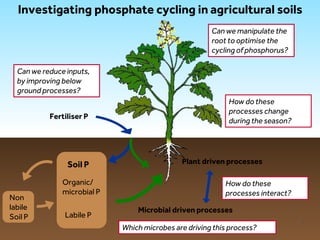
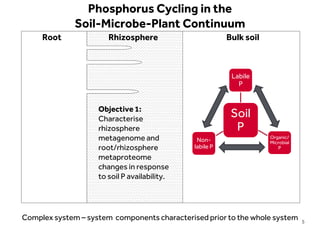
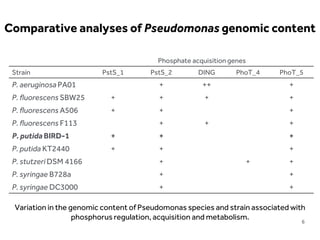
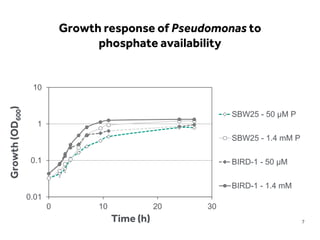
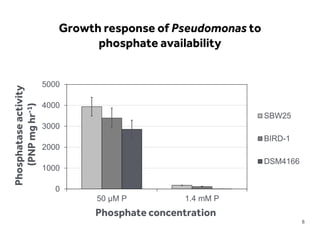
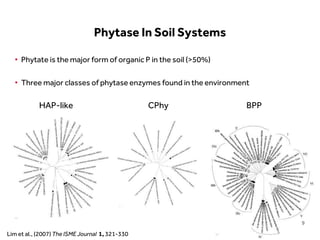
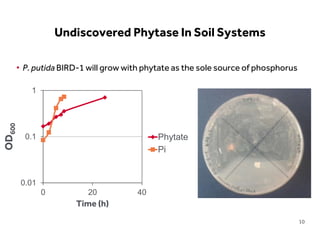
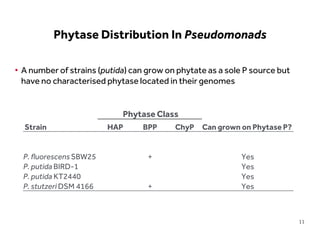
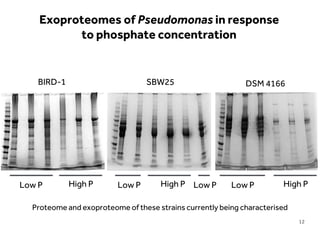
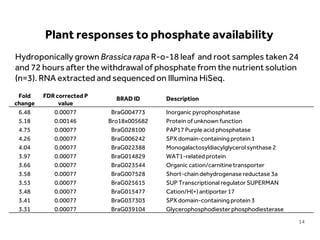
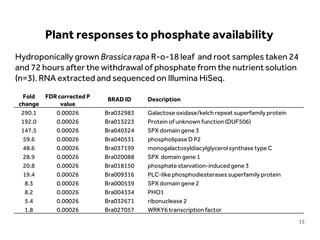
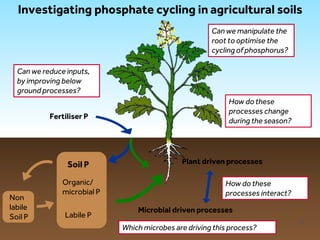
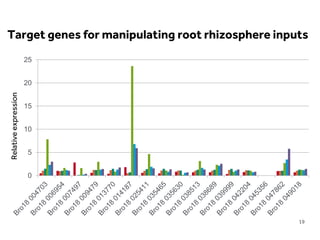
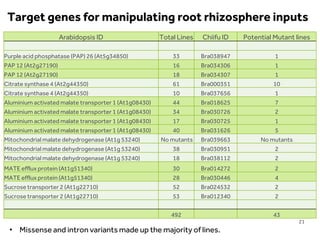
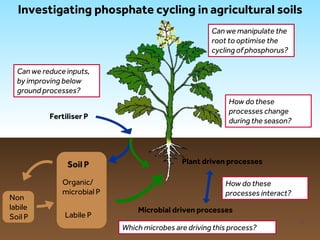
This document discusses research on phosphorus cycling in the soil-microbe-plant system. It summarizes work characterizing components like soil bacteria genomes and plant responses to phosphorus availability. The research aims to investigate phosphate cycling over time in agricultural soils by studying plant, microbial and root gene expression changes. It has obtained plant mutants targeted at genes influencing rhizosphere inputs to manipulate below-ground phosphorus cycling.