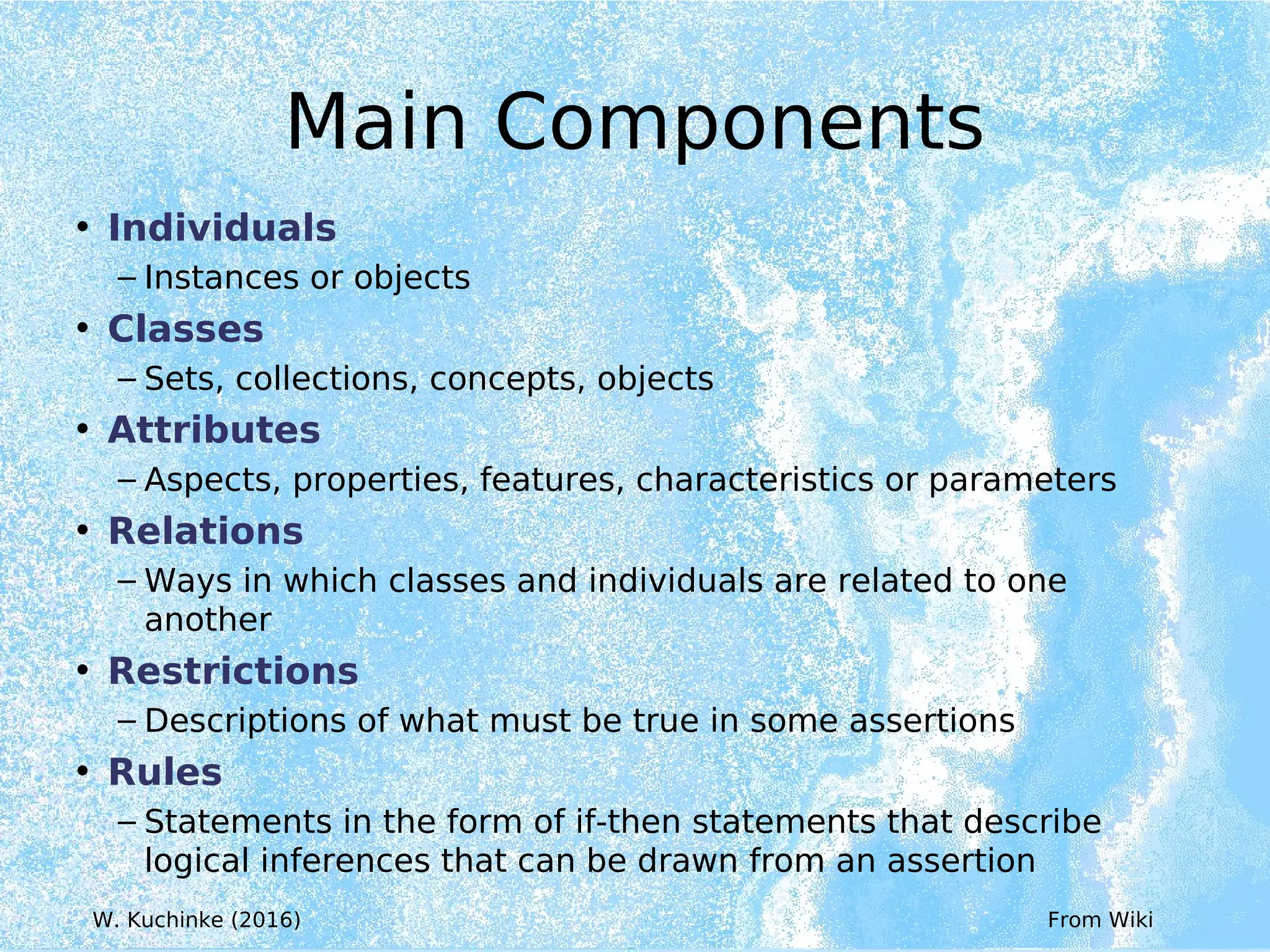
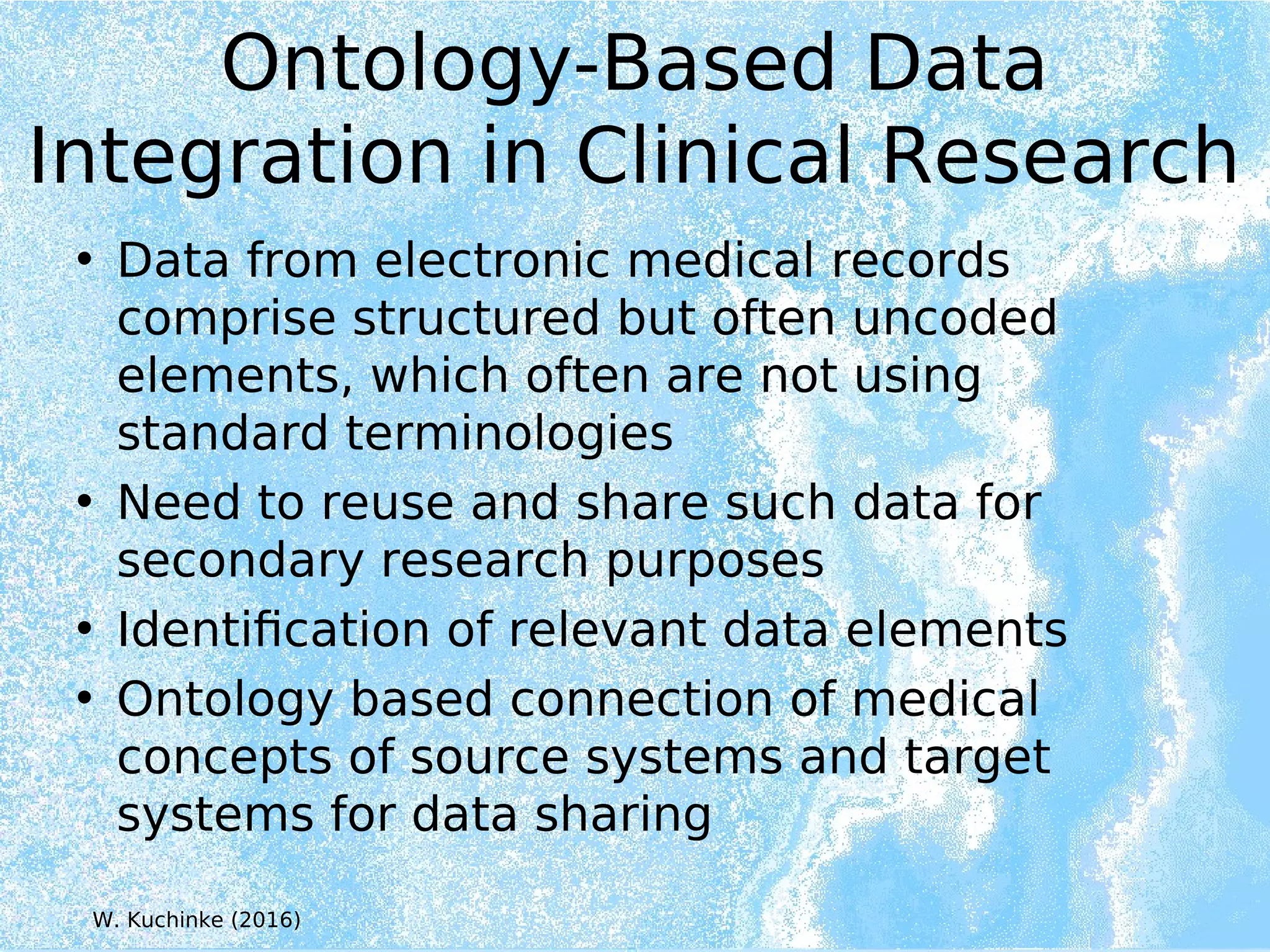
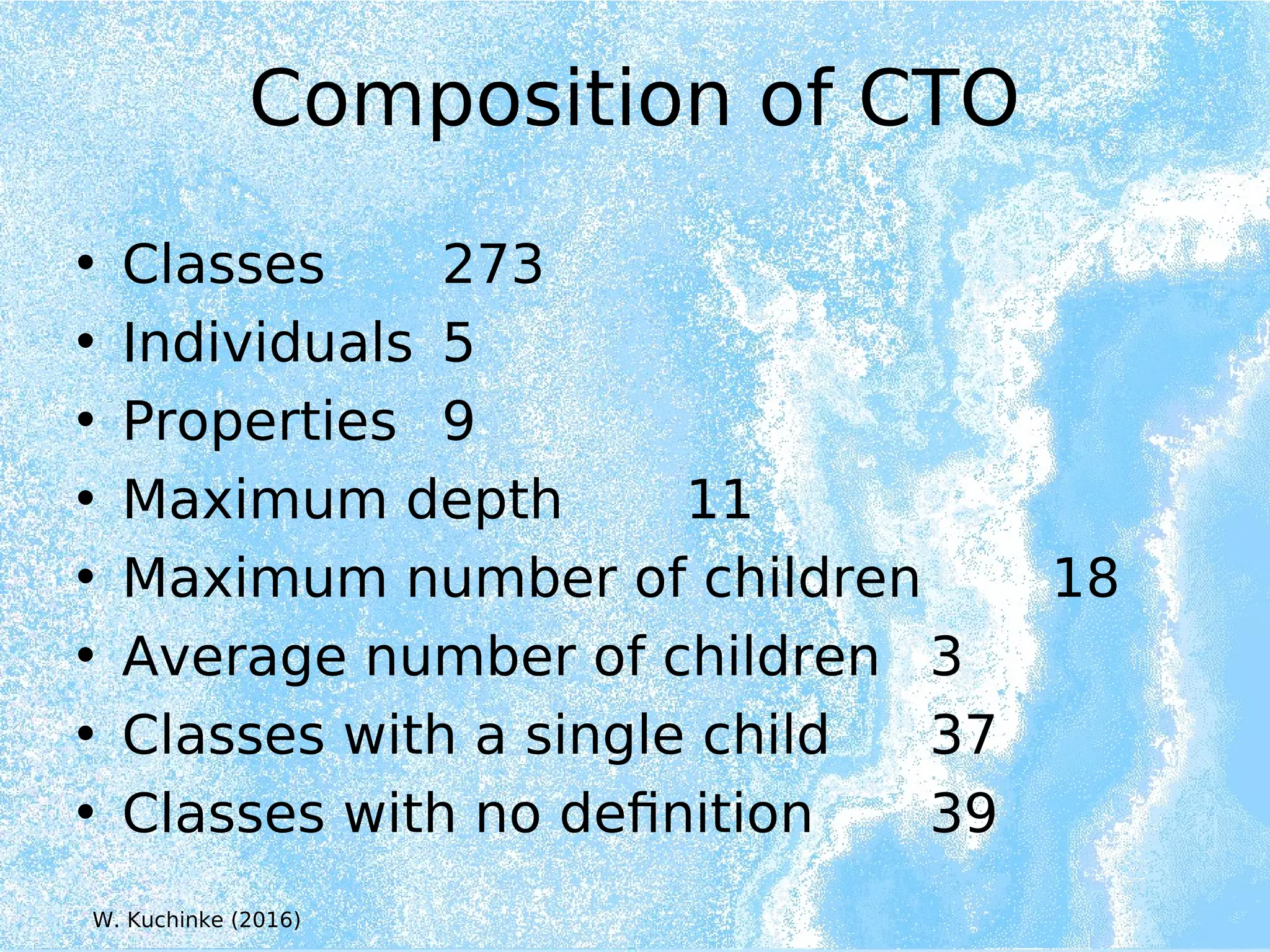
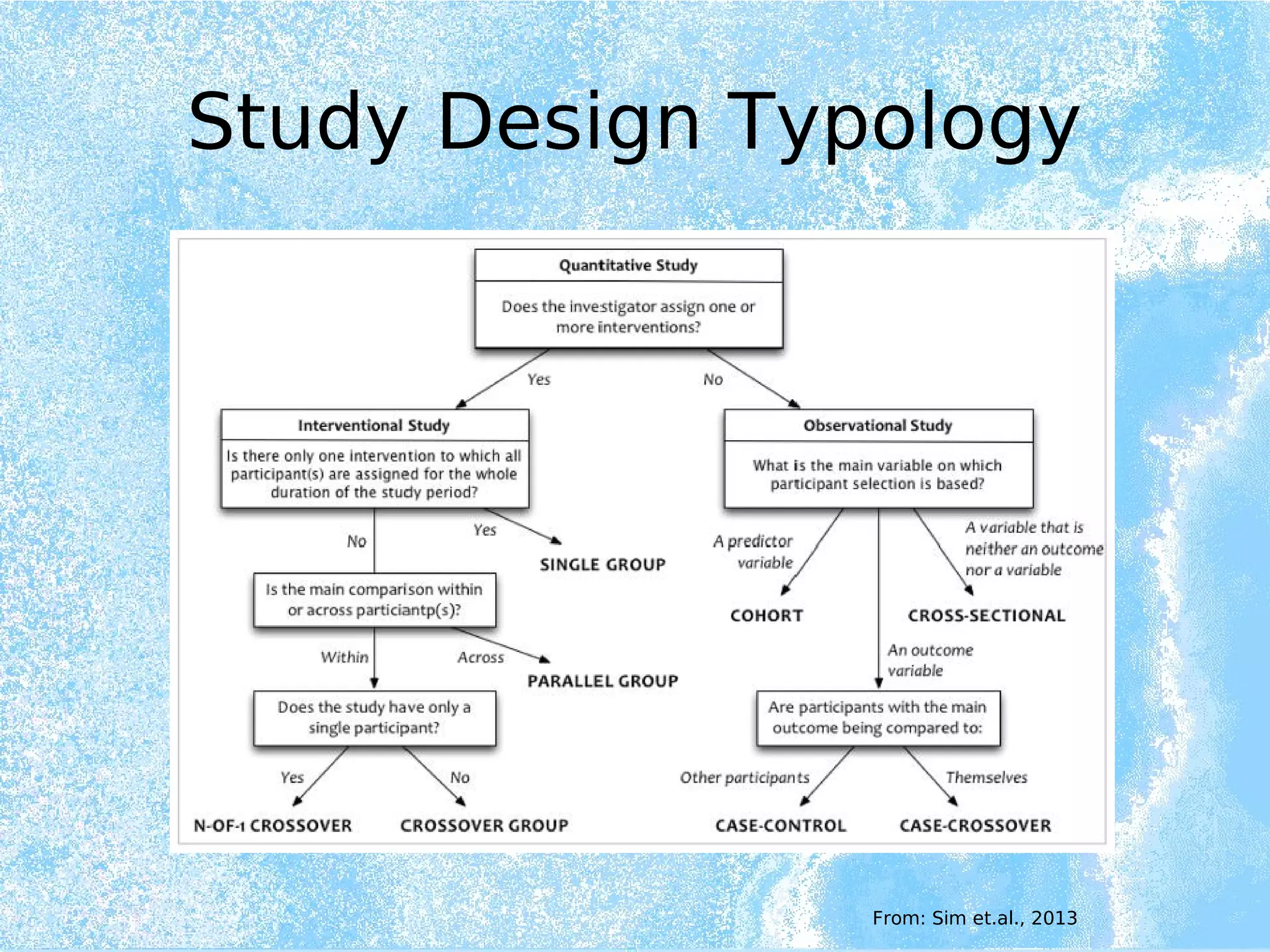
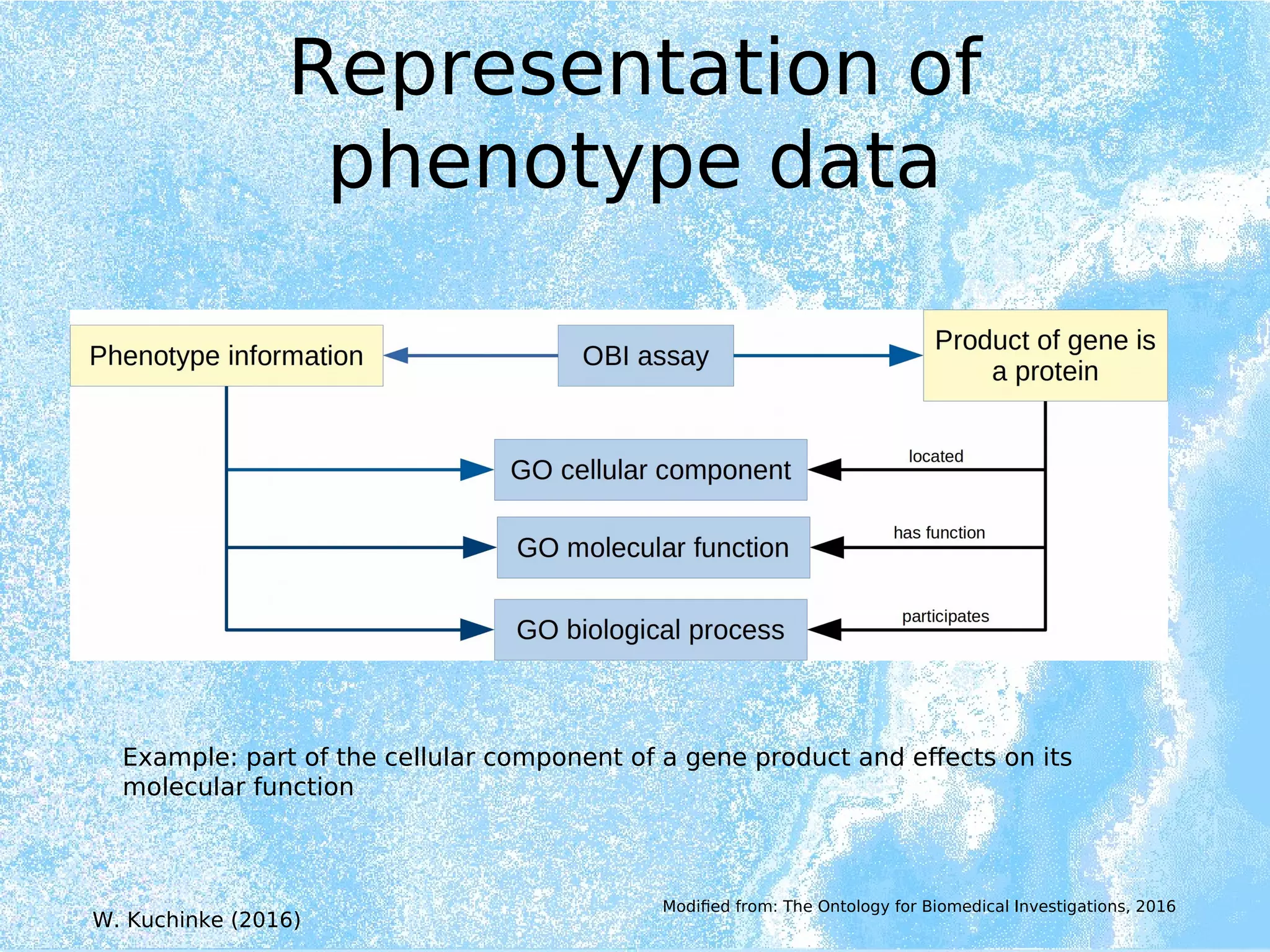
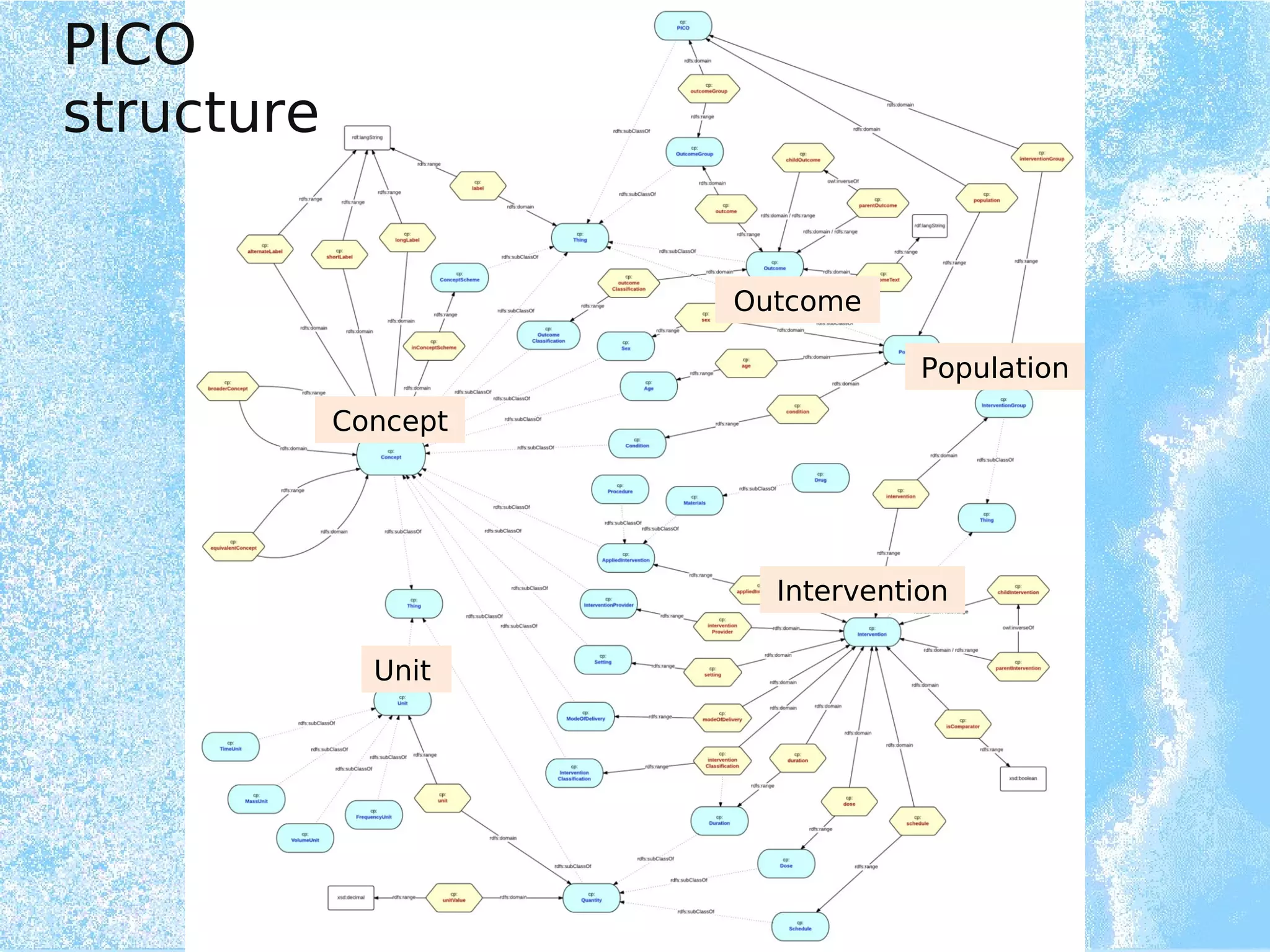
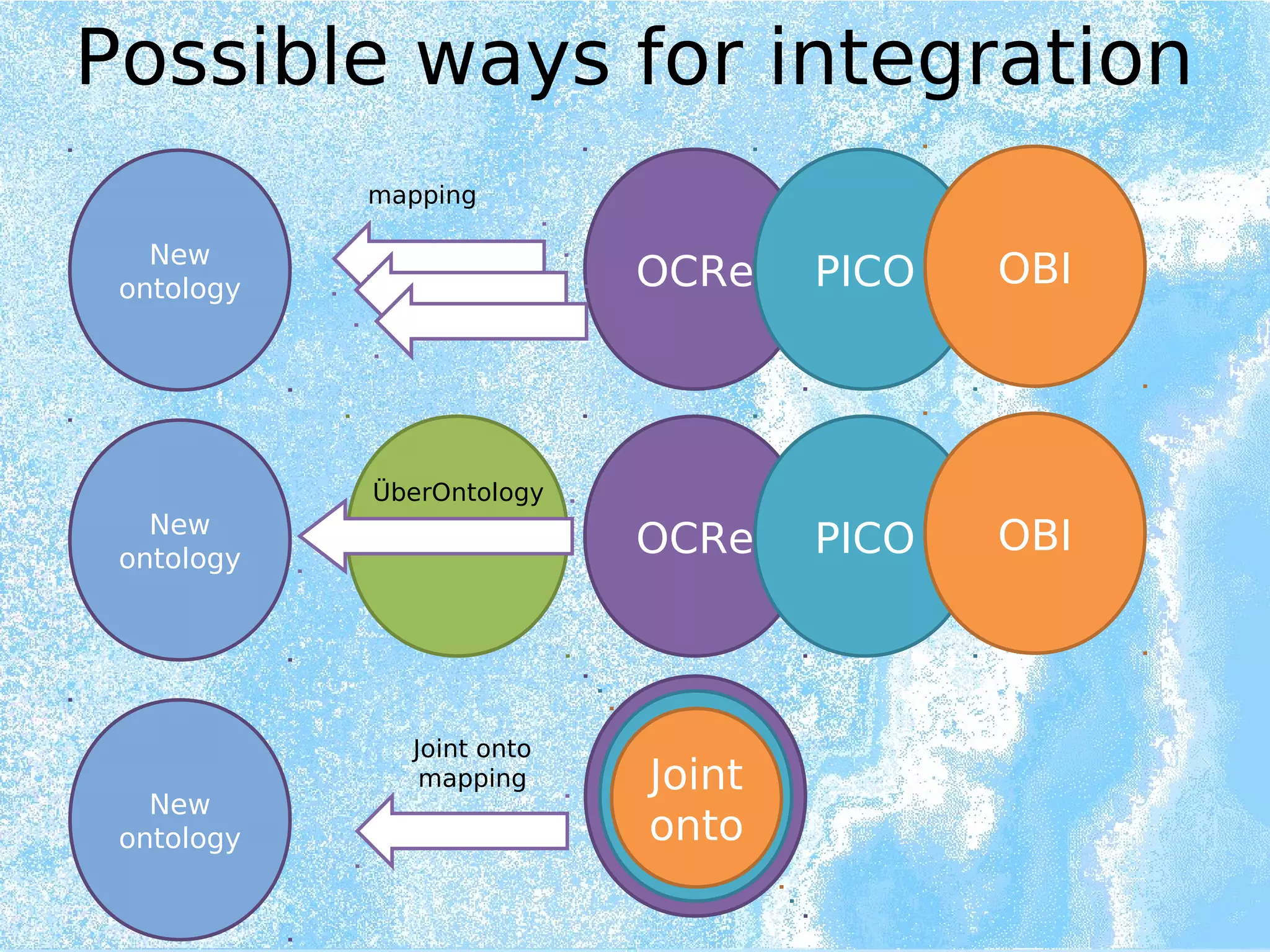
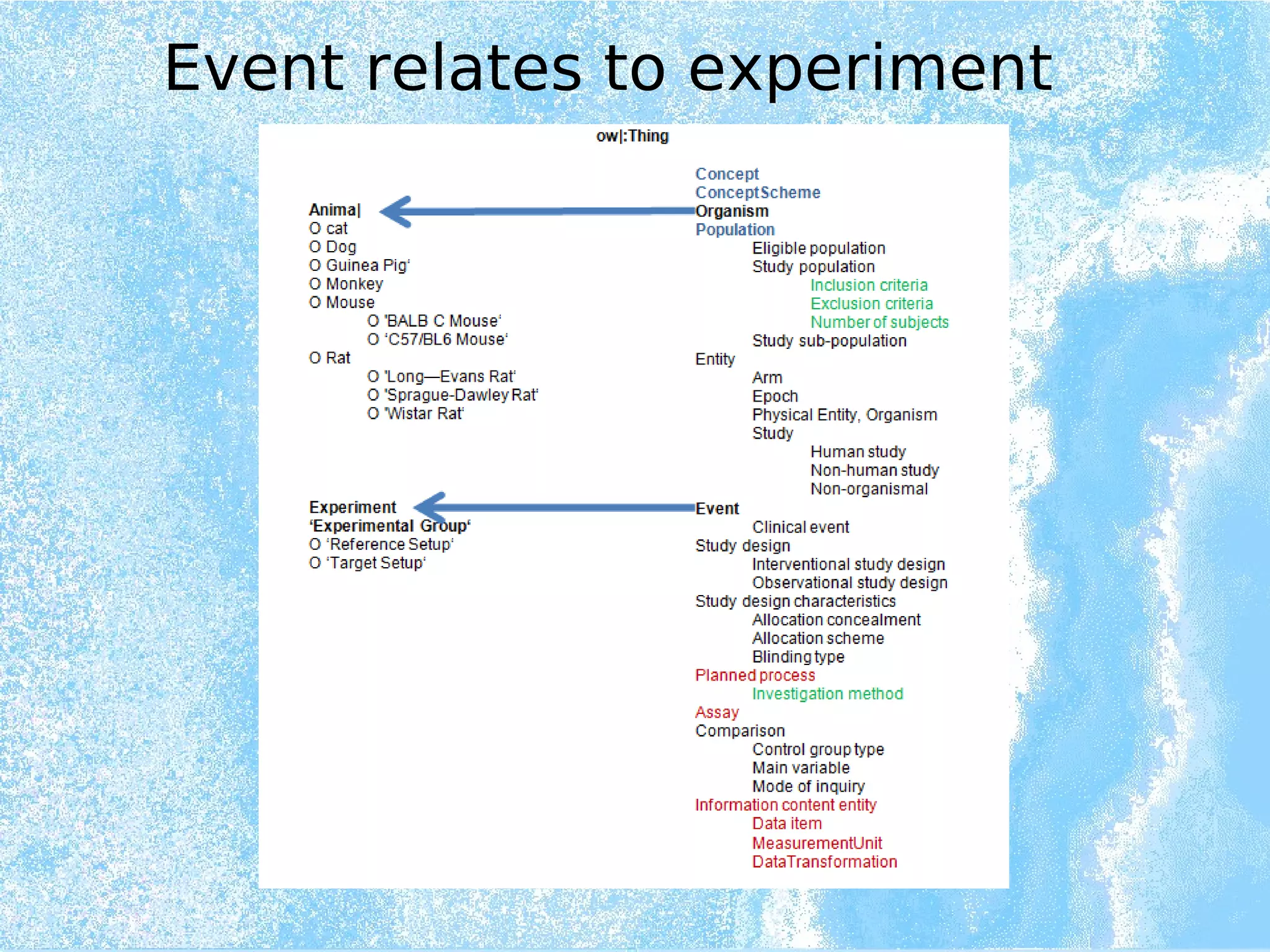
Wolfgang Kuchinke presented on ontologies for clinical research. He discussed what ontologies are and their main components. Their purpose is to limit complexity and organize data into information and knowledge. Kuchinke described several existing ontologies for clinical research including the Clinical Trial Ontology, Ontology of Clinical Research, Ontology for Biomedical Investigations, and Cochrane PICO Ontology. He noted the need for an ontology to integrate clinical trial data and discussed possible ways to build a new joint ontology combining aspects of existing ones like PICO, OCRe, and OBI to better enable data reuse and sharing in clinical research.