NUCLEOTIDES(1).pptx Presentation on nucleotides structure
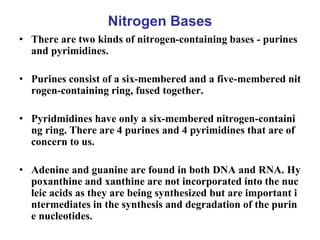
- 1. Nitrogen Bases • There are two kinds of nitrogen-containing bases - purines and pyrimidines. • Purines consist of a six-membered and a five-membered nit rogen-containing ring, fused together. • Pyridmidines have only a six-membered nitrogen-containi ng ring. There are 4 purines and 4 pyrimidines that are of concern to us. • Adenine and guanine are found in both DNA and RNA. Hy poxanthine and xanthine are not incorporated into the nuc leic acids as they are being synthesized but are important i ntermediates in the synthesis and degradation of the purin e nucleotides.
- 2. Purines
- 3. Pyrimidines
- 4. Nucleotides • A nucleotide is a chemical compound that consists of 3 p ortions: a nitrogenous base, a sugar, and one or more ph osphate groups • In the most common nucleotides the base is a derivative of purine or pyrimidine, and the sugar is the pentose deo xyribose or ribose. • Nucleotides are the structural units of RNA, DNA, and s everal cofactors - CoA, flavin adenine dinucleotide, flavi n mononucleotide, adenosine triphosphate and nicotina mide adenine dinucleotide phosphate. In the cell they ha ve important roles in metabolism and signaling.
- 5. Components of Nucleotides • Nitrogenous bases: There are two kinds of nitrogen-cont aining bases - purines and pyrimidines. • Purines consist of a six-membered and a five-membered nitrogen-containing ring, fused together. • Pyridmidines have only a six-membered nitrogen-contai ning ring. • There are 4 purines and 4 pyrimidines that are of concer n to us.
- 6. Purines 1. Adenine = 6-amino purine 2. Guanine = 2-amino-6-oxy purine
- 7. Pyrimidines 1. Uracil = 2,4-dioxy pyrimidine 2. Thymine = 2,4-dioxy-5-methyl pyrimidine 3. Cytosine = 2-oxy-4-amino pyrimidine URACIL
- 8. Nucleosides • If a sugar, either ribose or 2-deoxyribose, is add ed to a nitrogen base, the resulting compound i s called a nucleoside. Carbon 1 of the sugar is a ttached to nitrogen 9 of a purine base or to nitro gen 1 of a pyrimidine base. • The names of purine nucleosides end in -osine and the names of pyrimidine nucleosides end in -idine. The convention is to number the ring ato ms of the base normally and to use l', etc. to dis tinguish the ring atoms of the sugar. • Unless otherwise specified, the sugar is assum ed to be ribose. To indicate that the sugar is 2'- deoxyribose, a d- is placed before the name.
- 15. Phosphoric Acid
- 16. Nucleotides
- 19. THE STRUCTURE OF DNA
- 20. Major forms of DNA • Three major forms – B-DNA – A-DNA – Z-DNA
- 21. A-DNA • Right-handed helix • Wider and flatter than b-DNA • 11.6 bp per turn • Pitch of 34 A • Base planes are tilted 20 degrees with respect to h elical axis – Helix axis passes “above” major groove – Deep major and shallow minor groove
- 22. • Observed under dehydrating conditions • When relative humidity is ~ 75% – B-DNA A-DNA (REVERSIBLE) • Most self-complementary oligonucleotides of < 10 bp crystallize in A-DNA configuration. • A-DNA has been observed in 2 contexts: – At active site of DNA polymerase (~ 3 bp ) – Gram (+) bacteria undergoing sporulation –Cross-linking of pyrimidine bases
- 23. Z-DNA • A left-handed helix • Seen in conditions of high salt concentrations – Reduces repulsions between closest phosphate grou ps on opposite strands (8 a vs 12 a in B-DNA) • In complementary polynucleotides with alternating purin es and pyrimidines – Poly d(GC) · poly d(GC) – Poly d(AC) poly d(GT) • Might also be seen in DNA segments with above chara cteristics
- 24. • 12 base pairs per turn • A pitch of 44 degrees • A deep minor groove • No discernible major groove • Reversible change from B-DNA to Z-DNA in loca lized regions may act as a “switch” to regulate gene expression
- 25. STRUCTURAL VARIANTS OF DNA • DEPEND UPON: – SOLVENT COMPOSITION • WATER • IONS – BASE COMPOSITION • IN-CLASS QUESTION: WHAT FORM OF DNA WOULD YOU EXPECT TO SEE IN DESSICATED BRINE SHRIMP EGGS? W HY?
- 26. • The chromosomes contain 90% of the cell ’s DNA. • 10% is present in mitochondria and chlorop lasts.
- 27. Differences between DNA and RNA • DNA is double stranded; RNA is a single stranded • DNA contains the pentose sugar deoxyribose; RNA contains the pentose sugar ribose. • DNA has the base Thymine (T) but not Uracil (U); RNA has U but not T. • DNA is very long (billions of bases); RNA is smaller (hundreds to thousands of bases) • DNA is self-replicating, RNA is copied from the DNA so it is no t self-replicating
- 28. The Structure of DNA
- 29. • DNA is a long polymer made from repeating units called nucleo tides. • The DNA chain is 22 to 26 Ångströms wide (2.2 to 2.6 nanomet res), and one nucleotide unit is 3.3 Å (0.33 nm) long. • Although each individual repeating unit is very small, DNA pol ymers can be enormous molecules containing millions of nucle otides. • DNA does not usually exist as a single molecule, but instead as a tightly-associated pair of molecules.
- 30. • These two long strands entwine like vines, in the shape of a double helix. • In double stranded linear DNA, 1 end of each strand has a fr ee 5’ carbon and 1 end has a free 3’ OH group. • The two strands are in the opposite orientation with respect to each other (antiparallel). • Adenines always base pair with thymines (2 hydrogen bonds ) and guanines always base pair with cytosines (3 hydrogen bonds); this is called the Chargaff’s rule.
- 32. DNA Double Helix P P P O O O 1 2 3 4 5 5 3 3 5 P P P O O O 1 2 3 4 5 5 3 5 3 G C T A
- 33. • The backbone of the DNA strand is made from alter nating phosphate and sugar residues. • The sugar in DNA is 2-deoxyribose, which is a pento se (five-carbon) sugar. The sugars are joined togethe r by phosphate groups that form phosphodiester bon ds between the third and fifth carbon atoms of adjac ent sugar rings. • In a double helix the direction of the nucleotides in o ne strand is opposite to their direction in the other st rand. This arrangement of DNA strands is called ant iparallel. The asymmetric ends of DNA strands are r eferred to as the 5′ (five prime) and 3′ (three prime) e nds.
- 34. • The DNA double helix is stabilized by hydrogen bon ds between the bases attached to the two strands. Th e four bases found in DNA are adenine (A), cytosine (C), guanine (G) and thymine (T). • These four bases are attached to the sugar and a pho sphate to form the complete nucleotide, as shown for adenosine monophosphate. • The double helix is a right-handed spiral. The DNA strands wind around each other leaving gaps betwee n each set of phosphate backbones.
- 35. • There are two of these grooves twisting around the surfa ce of the double helix: the major groove, is 22 Å wide an d the minor groove, is 12 Å wide. • Each type of base on one strand forms a bond with just o ne type of base on the other strand. This is called comple mentary base pairing. Here, purines form hydrogen bon ds to pyrimidines, with A bonding only to T, and C bond ing only to G. • The double helix is also stabilized by the hydrophobic eff ect and pi stacking, which are not influenced by the sequ ence of the DNA. • The two strands of DNA in a double helix can be pulled apart either by a mechanical force or high temperature.
- 36. RNA • UNLIKE DNA, RNA IS SYNTHESIZED AS A SINGLE STRAND • THERE ARE DOUBLE-STRANDED RNA STRUCTURES – RNA CAN FOLD BACK ON ITSELF – DEPENDS ON BASE SEQUENCE – GIVES STEM (DOUBLE-STRAND) AND LOOP (SINGLE-STRAND ST RUCTURES) • DS RNA HAS AN A-LIKE CONFORMATION – STERIC CLASHES BETWEEN 2’-OH GROUPS PREVENT THE B-LIK E CONFORMATION
- 37. DNA REPLICATION •DNA replication begins with the "unzipping" of the parent molecule as the hydrogen bonds between the b ase pairs are broken by the helicase enzyme. • The new strands are assembled from deoxynucleosid e triphosphates that are added to short segment of R NA known as an RNA primer. The primer is "laid do wn" complementary to the DNA template by an enzy me known as RNA polymerase or Primase.
- 38. • Each incoming nucleotide is covalently linke d to the "free" 3' carbon atom on the pentos e as the second and third phosphates are re moved as a molecule of pyrophosphate (PPi) . • The nucleotides are assembled complementa ry to the order of bases on the strand servin g as the template
- 40. Replication Fork
- 42. The Enzymes DNA synthesis • A portion of the double helix is unwound by a helicase. • A molecule of a DNA polymerase binds to one strand of the DN A and begins moving along it in the 3' to 5' direction, using it as a template for assembling a leading strand of nucleotides and re forming a double helix. In eukaryotes, this molecule is called D NA polymerase delta (δ). • Because DNA synthesis can only occur 5' to 3', a molecule of a s econd type of DNA polymerase (epsilon, ε, in eukaryotes) binds to the other template strand as the double helix opens and adds dNTPs to RNA Primers laid on the lagging strand by the RNA Primase. • This molecule synthesizes discontinuous segments of polynucleo tides called Okazaki fragments.
- 43. • Exonuclease activity of DNA Polymerase I Finds and remo ves the RNA Primers • Another enzyme, DNA ligase I then stitches the Okazaki fr agments together in the lagging strand. • Termination of DNA replication happens when the DNA P olymerase reaches to an end of the strands. • The end of the parental strand where the last primer binds isn't replicated. These ends of linear (chromosomal) DNA c onsists of non-coding DNA that contains repeat sequences and are called telomeres. As a result, a part of the telomere is removed in every cycle of DNA Replication. • Nucleases remove wrong nucleotides from the daughter str and.
- 44. DNA damage and repair
- 45. DNA damage Damage caused by exogenous agents • UV-B light causes crosslinking between adjacent cytosi ne and thymine bases creating pyrimidine dimers. This is called direct DNA damage. • UV-A light creates mostly free radicals - especially if s unscreen penetrated into the skin. The damage caused by free radicals is called indirect DNA damage. • Ionizing radiation such as that created by radioactive decay or in cosmic rays causes breaks in DNA strands. • Thermal disruption at elevated temperature increases t he rate of depurination (loss of purine bases from the DNA backbone) and single strand breaks.
- 46. • Industrial chemicals such as vinyl chloride and hydroge n peroxide, and environmental chemicals such as polyc yclic hydrocarbons found in smoke, soot and tar create a huge diversity of DNA adducts- ethenobases, oxidize d bases, alkylated phosphotriesters and Cross linking o f DNA just to name a few. • UV damage, alkylation/methylation, X-ray damage an d oxidative damage are examples of induced damage. S pontaneous damage can include the loss of a base, dea mination, sugar ring puckering and tautomeric shift.
- 47. DNA damage due to endogenous cellular processes • There are four main types: • oxidation of bases [e.g. 8-oxo-7,8-dihydroguanine (8-ox oG)] and generation of DNA strand interruptions fro m reactive oxygen species, • alkylation of bases (usually methylation), such as form ation of 7-methylguanine, 1-methyladenine, O6 methyl guanine • hydrolysis of bases, such as deamination, depurination and depyrimidination. • mismatch of bases, due to errors in DNA replication, in which the wrong DNA base is stitched into place in a n ewly forming DNA strand, or a DNA base is skipped o ver or mistakenly inserted.
- 49. Direct reversal • Cells are known to eliminate three types of damage to their DN A by chemically reversing it. These mechanisms do not require a template, since the types of damage they counteract can only o ccur in one of the four bases. • Such direct reversal mechanisms are specific to the type of da mage incurred and do not involve breakage of the phosphodiest er backbone. • The formation of thymine dimers (a common type of cyclobutyl dimer) upon irradiation with UV light results in an abnormal c ovalent bond between adjacent thymidine bases. • The photoreactivation process directly reverses this damage by the action of the enzyme photolyase, whose activation is obligate ly dependent on energy absorbed from blue/UV light (300–500n m wavelength) to promote catalysis.
- 50. • Another type of damage, methylation of guanine bases, is directly reversed by the protein methyl guanine met hyl transferase (MGMT), the bacterial equivalent of w hich is called as ogt. • This is an expensive process because each MGMT mol ecule can only be used once; that is, the reaction is stoi chiometric rather than catalytic. • A generalized response to methylating agents in bacter ia is known as the adaptive response and confers a lev el of resistance to alkylating agents upon sustained exp osure by upregulation of alkylation repair enzymes. • The third type of DNA damage reversed by cells is cer tain methylation of the bases cytosine and adenine.
- 51. • The base-excision repair enzyme is uracil-DNA glycosyl ase. When only one of the two strands of a double helix has a defect, the other strand can be used as a template to guide the correction of the damaged strand. The excision repair mechanisms will remove the da maged nucleotide and replace it with an undamaged nucleotide complementary to that found in the unda maged DNA strand are; • Base excision repair (BER), which repairs damage to a single base caused by oxidation, alkylation, hydrolysis, or deamination. The damaged base is removed by a DN A glycosylase, resynthesized by a DNA polymerase, and a DNA ligase performs the final nick-sealing step. Single strand damage
- 52. • Nucleotide excision repair (NER), which recog nizes bulky, helix-distorting lesions such as pyr imidine dimers and 6,4 photoproducts. • A specialized form of NER known as transcript ion-coupled repair deploys NER enzymes to ge nes that are being actively transcribed. • Mismatch repair (MMR), which corrects error s of DNA replication and recombination that re sult in mispaired (but undamaged) nucleotides.
- 53. Double strand damage • Double-strand breaks (DSBs), in which both strands in the dou ble helix are severed, are particularly hazardous to the cell bec ause they can lead to genome rearrangements. Two mechanism s exist to repair DSBs: non-homologous end joining (NHEJ) an d recombinational repair (also known as template-assisted rep air or homologous recombination repair). • DNA ligase is the enzyme that joins broken nucleotides togethe r by catalyzing the formation of an internucleotide ester bond between the phosphate backbone and the deoxyribose nucleoti des. • In NHEJ, DNA Ligase IV, a specialized DNA Ligase that forms a complex with the cofactor XRCC4, directly joins the two end s. To guide accurate repair, NHEJ relies on short homologous s equences called microhomologies present on the single-strande d tails of the DNA ends to be joined. If these overhangs are co mpatible, repair is usually accurate
- 54. Repair of damaged bases • DNA's bases may be modified by deamination or alk ylation. The position of the modified (damaged) bas e is called the "abasic site" or "AP site". • In E.coli, the DNA glycosylase can recognize the AP site and remove its base. • Then, the AP endonuclease removes the AP site and neighboring nucleotides. • The gap is filled by DNA polymerase I and DNA liga se.
- 55. DNA repair by excision
- 56. • In E. coli, proteins UvrA, UvrB, and UvrC are involve d in removing the damaged nucleotides (e.g., the dim er induced by UV light). • The gap is then filled by DNA polymerase I and DNA l igase. • In yeast, the proteins similar to Uvr's are named RA Dxx ("RAD" stands for "radiation"), such as RAD3, R AD10. etc.
- 58. Mismatch repair • To repair mismatched bases, the system has to know which bas e is the correct one. In E. coli, this is achieved by a special meth ylase called the "Dam methylase", which can methylate all aden ines that occur within (5')GATC sequences. • Immediately after DNA replication, the template strand has alr eady been methylated, but the newly synthesized strand is not methylated yet. Thus, the template strand and the new strand c an be distinguished. • The repairing process begins with the protein MutS which bind s to mismatched base pairs.
- 59. • Then, MutL is recruited to the complex and activates MutH w hich binds to GATC sequences. Activation of MutH cleaves th e unmethylated strand at the GATC site. • Subsequently, the segment from the cleavage site to the mismat ch is removed by exonuclease (with assistance from helicase II and SSB proteins). • If the cleavage occurs on the 3' side of the mismatch, this step i s carried out by exonuclease I (which degrades a single strand only in the 3' to 5' direction). • If the cleavage occurs on the 5' side of the mismatch, exonuclea se VII or RecJ is used to degrade the single stranded DNA. Th e gap is filled by DNA polymerase III and DNA ligase.
- 60. Mismatch repair
- 61. • The distance between the GATC site and the mis match could be as long as 1,000 base pairs. There fore, mismatch repair is very expensive and ineffi cient. • Mismatch repair in eukaryotes may be similar to that in E. coli. Homologs of MutS and MutL have been identified in yeast, mammals, and other euka ryotes. MSH1 to MSH5 are homologous to MutS; MLH1, PMS1 and PMS2 are homologous to Mut L. Mutations of MSH2, PMS1 and PMS2 are rela ted to colon cancer. • In eukaryotes, the mechanism to distinguish the te mplate strand from the new strand is still unclear.
- 62. Transcription •Prokaryotic transcription occurs in the cytoplasm alongside tra nslation. •Eukaryotic transcription is primarily localized to the nucleus. T he transcript is then transported into the cytoplasm where transl ation occurs. •Another important difference is that eukaryotic DNA is wound around histones to form nucleosomes and packaged as chromati n. Chromatin has a strong influence on the accessibility of the D NA to transcription factors and the transcriptional machinery in cluding RNA polymerase. •In prokaryotes, mRNA is not modified. Eukaryotic mRNA is m odified through RNA splicing, 5' end capping, and the addition o f a polyA tail.
- 63. Initiation •Transcription does not need a primer to start. RNA polymerase simply binds to theDNA and, along with ot her cofactors, unwinds the DNA to create an initiation bubble so that the RNA polymerase has access to the single-stranded DNA template. •In bacteria, transcription begins with the binding of RNA polymerase to the promoter in DNA. • The RNA polymerase is a core enzyme consisting of fiv e subunits: 2 α subunits, 1 β subunit, 1 β' subunit, and 1 ω subunit.
- 64. At the start of initiation, the core enzyme is associated with a sigma factor (number 70) that aids in finding the appropriate -35 and -10 basepairs downstream of promoter sequences. Simple diagram of transcription initiation. RNAP = RNA polymerase
- 65. Elongation • One strand of DNA, the template strand (or non-coding s trand), is used as a template for RNA synthesis. • As transcription proceeds, RNA polymerase traverses th e template strand and uses base pairing complementarit y with the DNA template to create an RNA copy. • Although RNA polymerase traverses the template stran d from 3' → 5', the coding (non-template) strand is usual ly used as the reference point, so transcription is said to go from 5' → 3'.
- 66. This produces an RNA molecule from 5' → 3', an exact copy of the coding strand (except that thymines are replaced with uracils. mRNA transcription can involve multiple RNA polymerases on a single DNA template and multiple rounds of replication. Simple diagram of transcription elongation
- 67. Termination • Bacteria use two different strategies for transcription te rmination: i) Rho-independent transcription termination where RN A transcription stops when the newly synthesized RNA molecule forms a hairpin loop, followed by a run of Us, which makes it detach from the DNA template. ii) "Rho-dependent" type of termination where a protein f actor called "Rho" destabilizes the interaction between the template and the mRNA, thus releasing the newly sy nthesized mRNA from the elongation complex.
- 68. Simple diagram of transcription termination
- 69. Post-transcriptional modifications of RNA • Post-transcriptional modification is a process by which, i n eukaryotic cells, the primary transcript RNA is convert ed into mature RNA e.g the conversion of precursor mess enger RNA into mature messenger RNA (mRNA) which i ncludes splicing and occurs prior to protein synthesis. • • This process is vital for the correct translation of the geno mes of eukaryotes as the human primary RNA transcript that is produced as a result of transcription contains both exons, which are coding sections of the primary RNA tran script and introns, which are the non coding sections of th e primary RNA transcript.
- 70. 5’ Capping • Capping involves the addition of 7-methylguanosine (m7G) to the 5' end. To achieve this, the terminal 5' ph osphate is removed, by the aid of a phosphatase enzy me. • Then enzyme guanosyl transferase catalyses the reacti on which produces the diphosphate 5' end. • The diphosphate 5' prime end then attacks the α phos phorus atom of a GTP molecule in order to add the gu anine residue in a 5'5' triphosphate link.
- 71. • The enzyme Guanine-7-methyltransferase then meth ylates the guanine ring at the N-7 position. • The ribose of the adjacent nucleotide may also be met hylated to give a cap 1. Methylation of nucleotides do wnstream of the RNA molecule produce cap 2, cap 3 s tructures and so on. In these cases the methyl groups are added to the 2' OH groups of the ribose sugar cata lyzed by 2’O-methyltransferase. • • The cap protects the 5' end of the primary RNA trans cript from attack by ribonucleases that have specificit y to the 3'5' phosphodiester bonds.
- 72. 3' Processing: Cleavage and Polyadenylation • The pre-mRNA processing at the 3' end of the RNA m olecule involves cleavage of its 3' end and then the add ition of about 200 adenine residues to form a poly(A) t ail. • The cleavage and adenylation reactions occur if a poly adenylation signal sequence (5'- AAUAAA-3') is locate d near the 3' end of the pre-mRNA molecule, which is followed by another sequence, which is usually (5'-CA- 3').
- 73. RNA Splicing • RNA splicing is the process by which introns, regions of RNA that do not code for protein, are removed from the pre-mRNA and the remaining exons connected to re-for m a single continuous molecule. • Although most RNA splicing occurs after the complete sy nthesis and end-capping of the pre-mRNA, transcripts wi th many exons can be spliced co-transcriptionally. • The splicing reaction is catalyzed by a large protein com plex called the spliceosome assembled from proteins and small nuclear RNA molecules that recognize splice sites i n the pre-mRNA sequence.
- 74. • Many pre-mRNAs, including those encoding antibod ies, can be spliced in multiple ways to produce differe nt mature mRNAs that encode different protein sequ ences. • This process is known as alternative splicing, and all ows production of a large variety of proteins from a l imited amount of DNA.
- 75. rRNA and tRNAs processing. • Ribosomal RNA are made from longer precursors ca lled preribosomal RNAs. In bacteria 16s,23s and 5s a rise from a 30s precursor while in eukaryotes 18s, 28 s and 5.8s RNAs are processed from a 45s preriboso mal RNA. 5s RNA in eukaryotes is made as a separat e transcript. • tRNA are derived from longer RNA precursors by e nzymatic removal of extra nucleotides from the 5’ an d 3’ ends.
- 76. • Addition of the CCA to the 3’ terminal end which is absent in some bacteria and all eukaryotes. • Modification of some bases by methylation, deami nation and reduction
- 77. Types of RNA Messenger RNA: • Messenger RNA (mRNA) is synthesized from a gene s egment of DNA which ultimately contains the informa tion on the primary sequence of amino acids in a prot ein to be synthesized. • The genetic code as translated is for m-RNA not DNA . The messenger RNA carries the code into the cytopla sm where protein synthesis occurs.
- 78. Ribosomal RNA • ribsomal RNA (rRNA) and protein combine to for m a nucleoprotein called a ribosome in the cytopla sm. • The ribosome serves as the site and carries the enz ymes necessary for protein synthesis. The ribosom e is made from two sub units, 50S and 30 S in prok aryotes and 60 and 40 in eukaryotes. • There are about equal parts rRNA and protein in a protein synthesising unit.
- 79. Ribosomal RNA • During protein synthesis, the ribosome attaches its elf to m-RNA and provides the stabilizing structur e to hold all substances in position as the pro tein is synthesized. • Several ribosomes may be attached to a sing le RNA at any time. In upper right corner is the 30S sub unit with mRNA and tRNA atta ched.
- 80. Transfer RNA • Transfer RNA (tRNA) contains about 75 nucleotides, thr ee of which are called anticodons, and one amino acid. T he tRNA reads the code and carries the amino acid to be incorporated into the developing protein. • There are at least 20 different tRNA's - one for each ami no acid. The basic structure of a tRNA is shown in the lef t graphic. • Part of the tRNA doubles back upon itself to form severa l double helical sections. On one end, the amino acid, the amino acid arm and on the opposite end, a specific base t riplet, called the anticodon arm, is used to actually "read " the codons on the mRNA.
- 82. • The 5'-terminal phosphate group. The acceptor stem i s a 7-base pair (bp) stem made by the base pairing of t he 5'-terminal nucleotide with the 3'-terminal nucleoti de. It contains the CCA 3'-terminal group used to atta ch the amino acid. • The acceptor stem may contain non-Watson-Crick ba se pairs. • The tail is a CCA sequence at the 3' end of the tRNA molecule. This sequence is important for the recogniti on of tRNA by enzymes critical in translation.
- 83. • In prokaryotes, the CCA sequence is transc ribed. In eukaryotes, the CCA sequence is a dded during processing and therefore does not appear in the tRNA gene.
- 84. • The D arm is a 4 bp stem ending in a loop that often c ontains dihydrouridine. • The anticodon arm is a 5-bp stem whose loop contains the anticodon. It also contains a Y that stands for a m odified purine nucleotide. • The T arm is a 5 bp stem containing the sequence TΨ C where Ψ is a pseudouridine. • Bases that have been modified, especially by methylati on, occur in several positions outside the anticodon. T he first anticodon base is sometimes modified to inosi ne or pseudouridine.
- 85. Transfer RNA • The tRNA "reads" the mRNA codon by using its own ant icodon. The actual "reading" is done by matching the bas e pairs through hydrogen bonding following the base pair ing principle. Each codon is "read" by various tRNA's un til the appropriate match of the anticodon with the codon occurs. • In this example, the tRNA anticodon (AAG) reads the cod on (UUC) on the mRNA. The UUC codon codes for pheny lalanine which is attached to the tRNA. Remember that t he codons read from the mRNA make up the genetic code as read by humans.
- 86. Small nuclear RNA (snRNA) • Small nuclear ribonucleic acid (snRNA) is a class of small RN A molecules that are found within the nucleus of eukaryotic ce lls. • They are transcribed by RNA polymerase II or RNA polymer ase III and are involved in a variety of important processes suc h as RNA splicing, regulation of transcription factors or RNA polymerase II, and maintaining the telomeres. • • They are always associated with specific proteins, and the com plexes are referred to as small nuclear ribonucleoproteins (sn RNP). These elements are rich in uridine content.
- 87. • A large group of snRNAs are known as small nucleol ar RNAs (snoRNAs). These are small RNA molecules that play an essential role in RNA biogenesis and gui de chemical modifications of ribosomal RNAs (rRNA s) and other RNA genes (tRNA and snRNAs). • They are located in the nucleolus and the Cajal bodie s of eukaryotic cells (the major sites of RNA synthesi s).