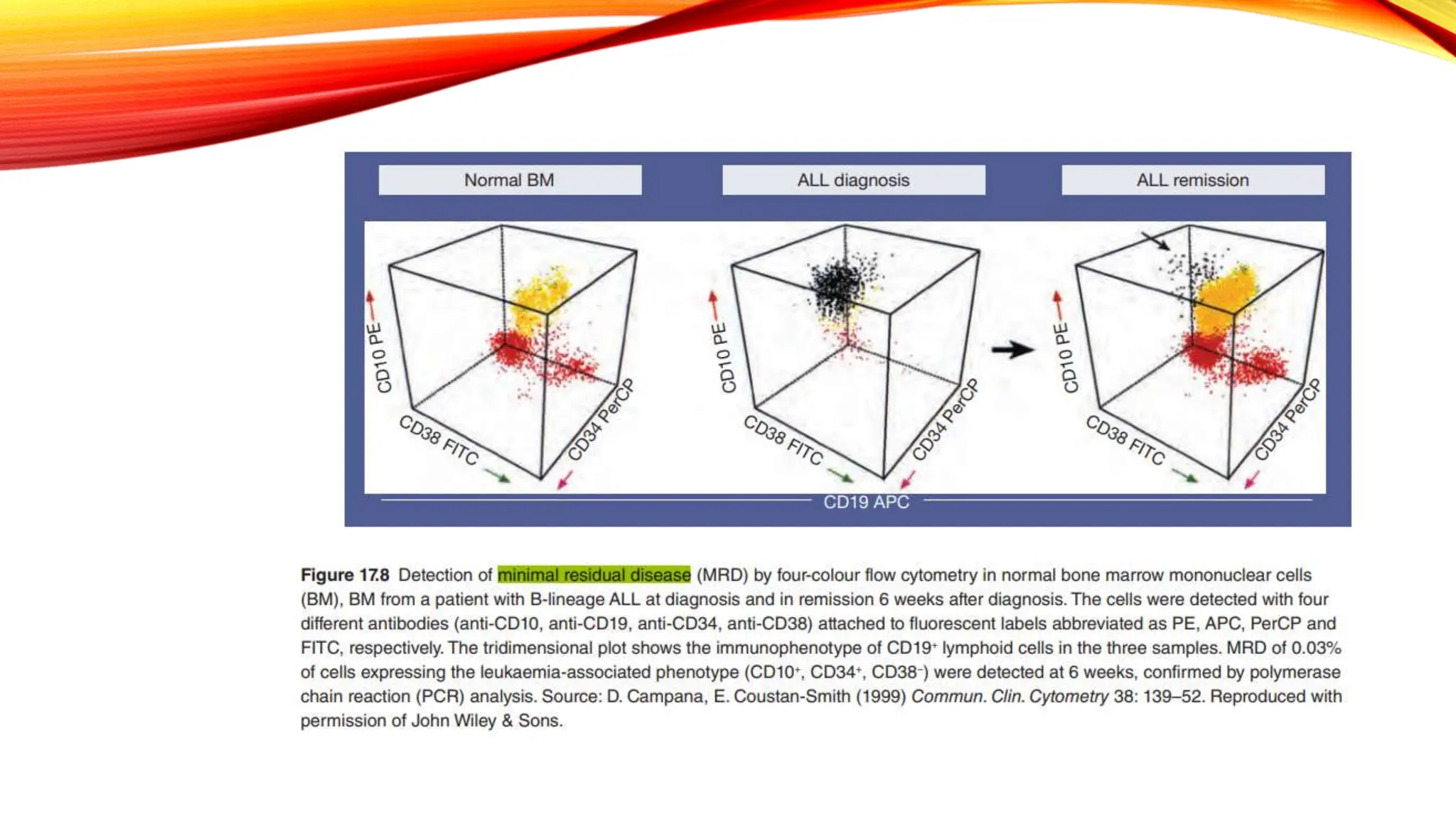
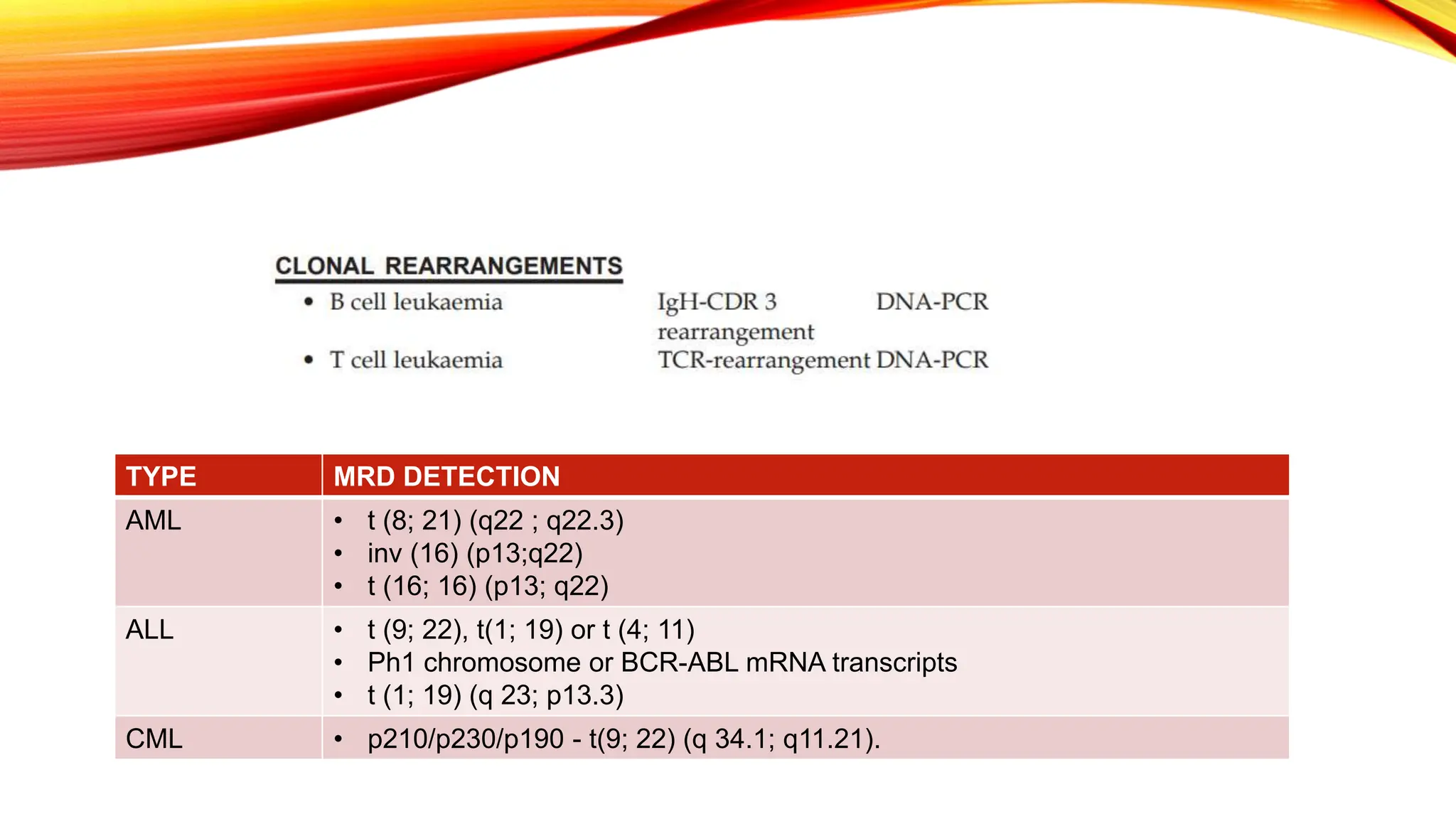
Minimal Residual Disease (MRD) refers to small quantities of cancer cells that persist after treatment, which can lead to relapse. MRD testing is crucial as it helps determine if additional treatment is needed and can predict treatment success and patient outcomes across various blood cancers. The document reviews various detection methods and emphasizes the importance of identifying and monitoring MRD for better cancer management.