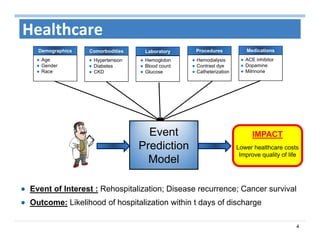
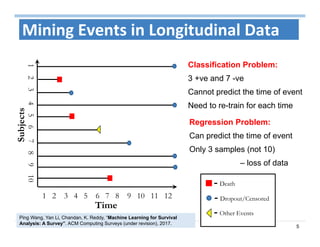
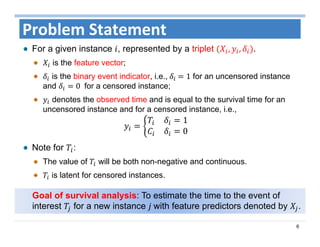
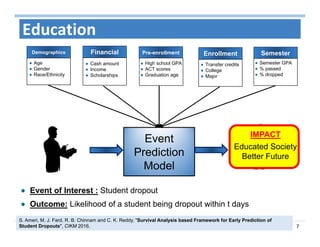
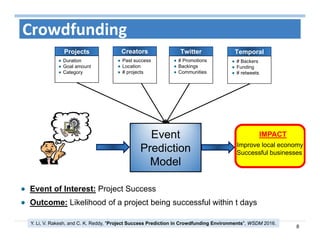
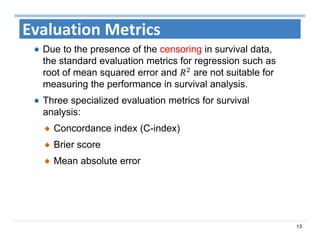
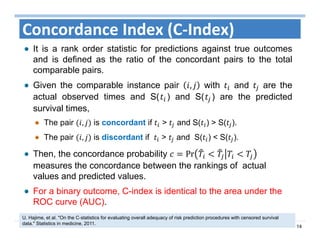
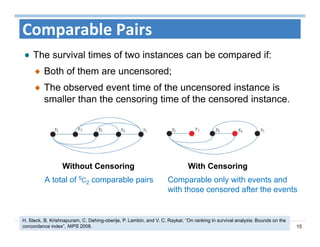
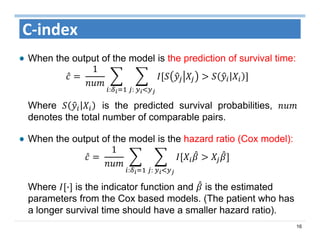
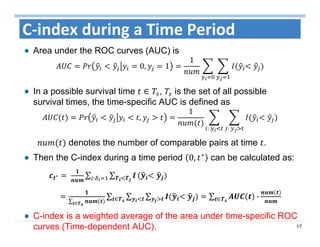
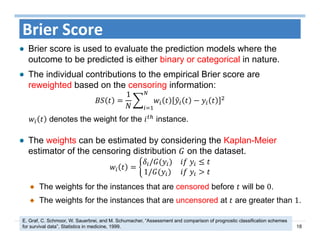
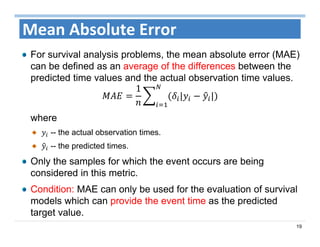
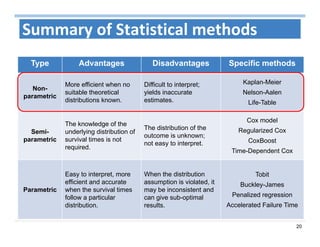
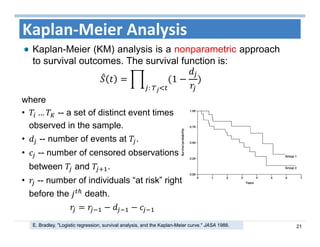
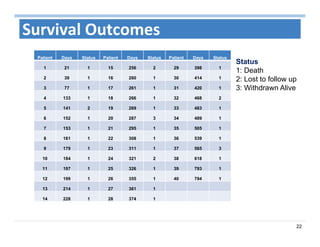
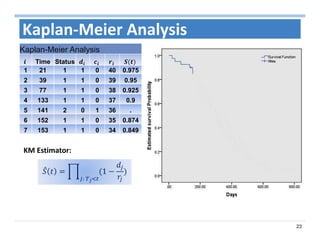
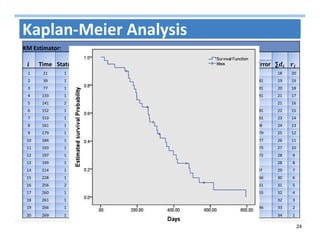
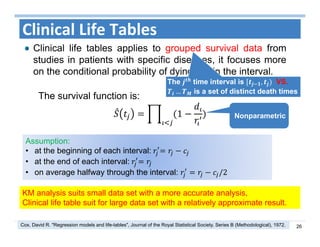
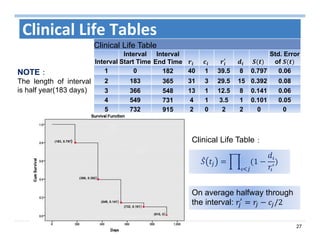
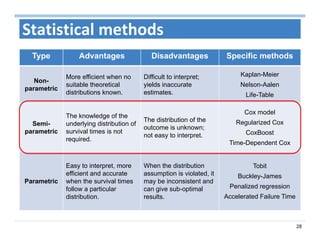
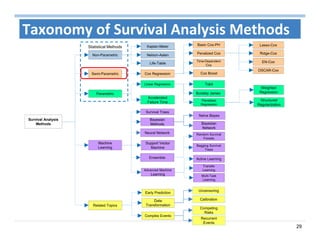
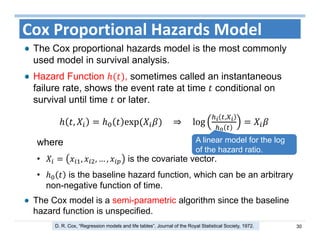
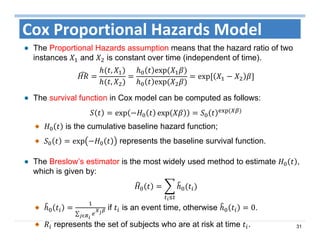
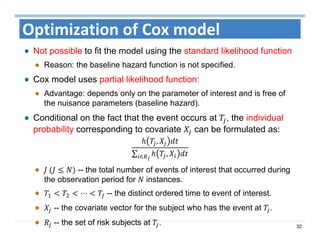
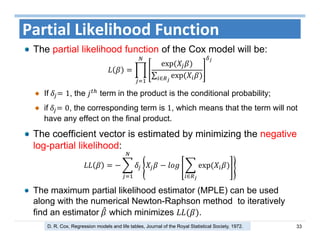
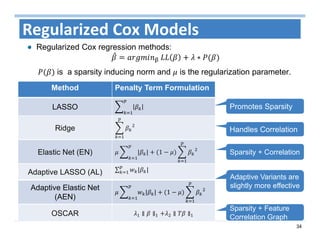
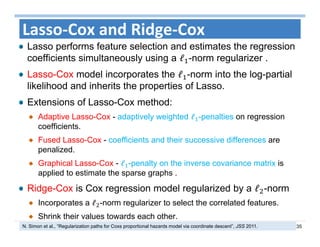
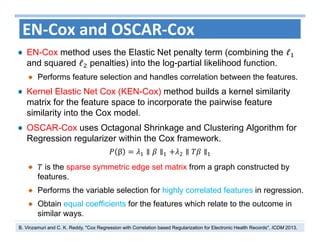
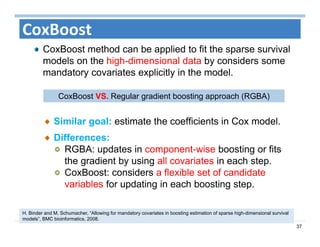
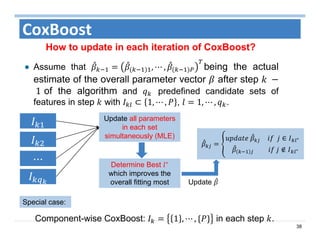
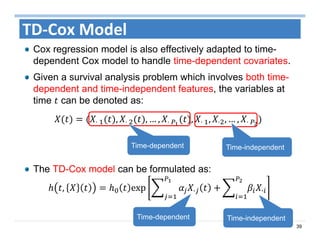
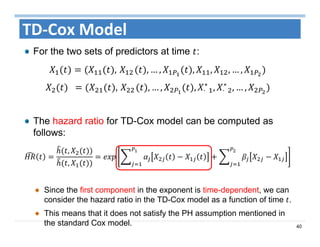
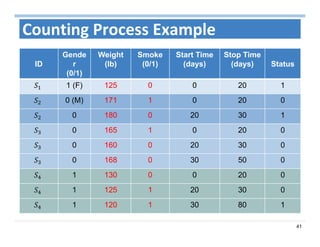
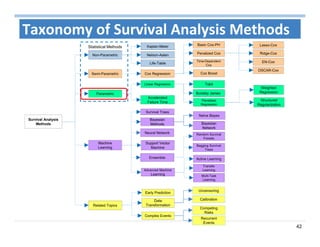
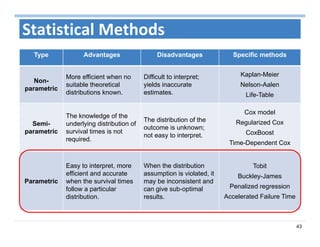
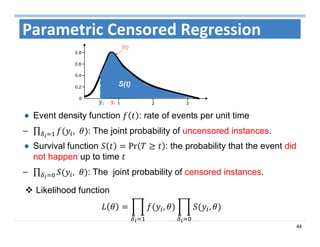
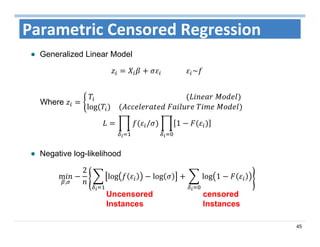
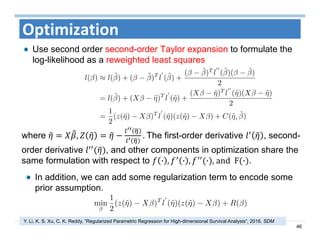
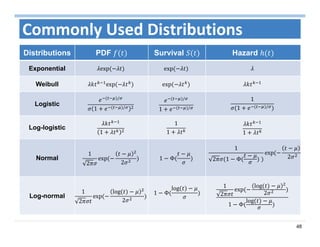
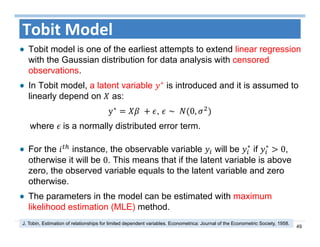
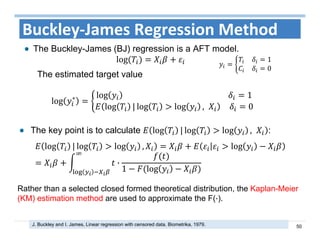
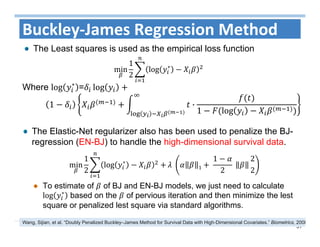
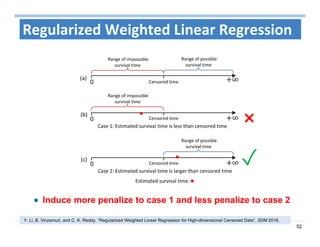
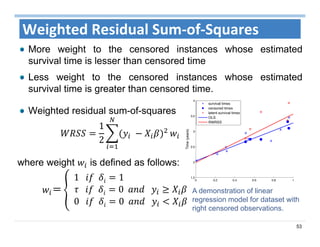
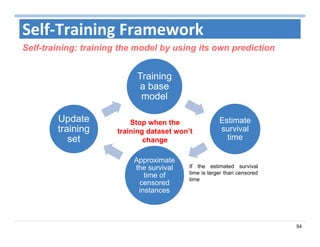
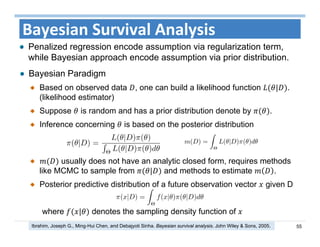
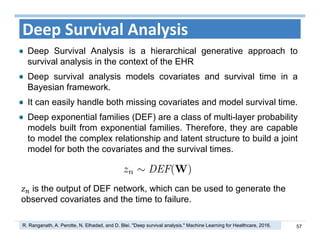
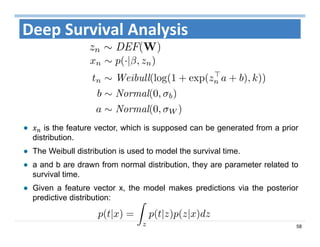
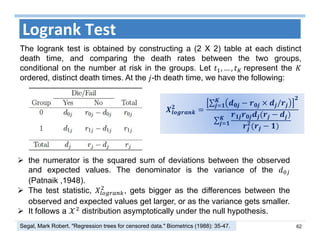
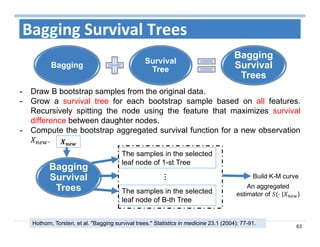
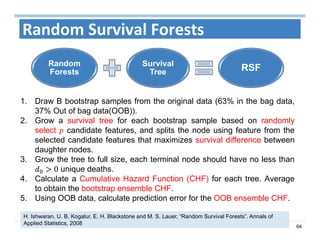
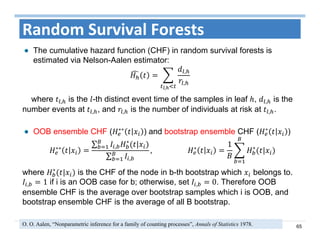
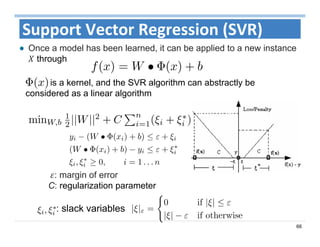
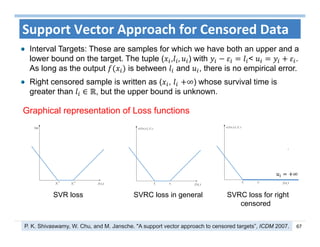
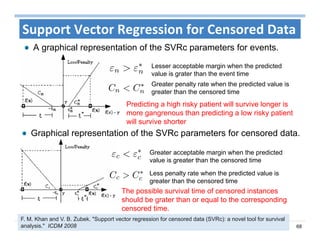
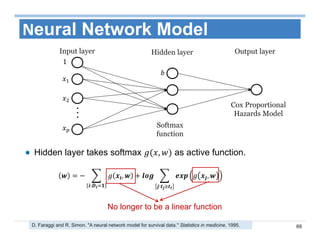
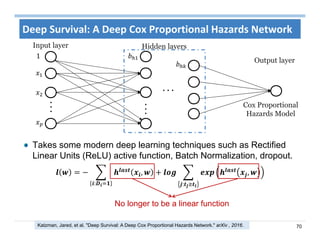
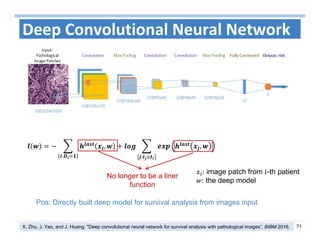
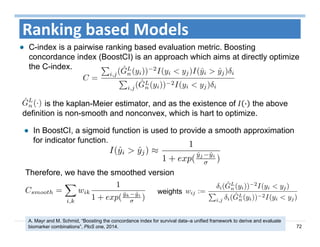
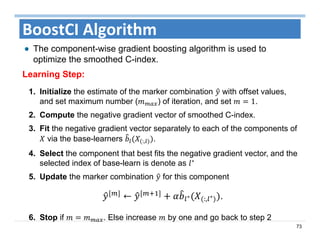
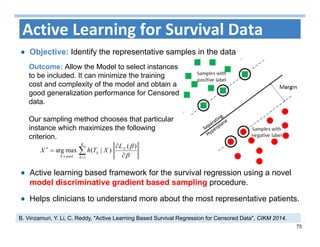
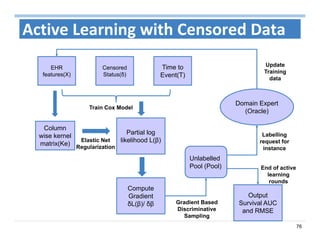
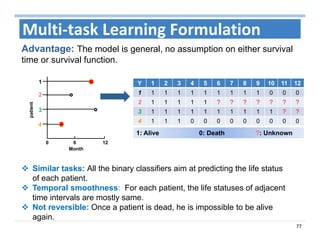
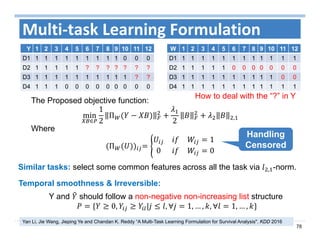
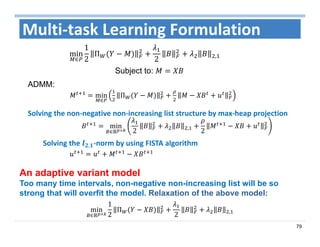
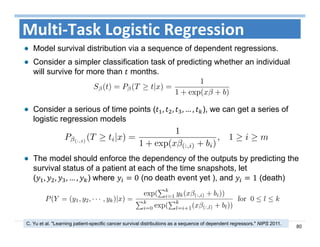
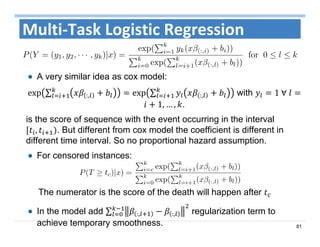
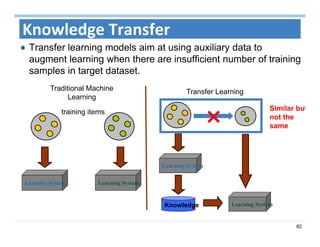
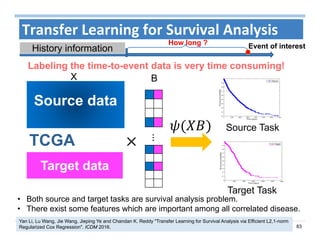
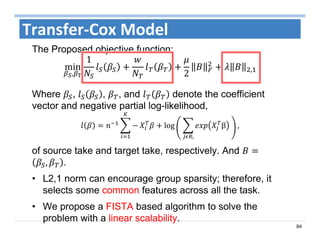
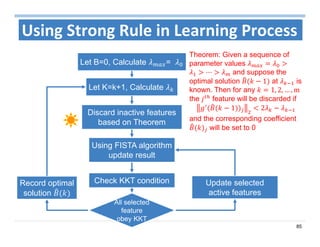
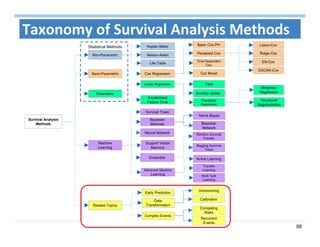
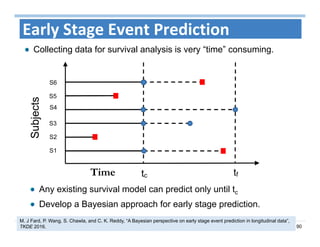
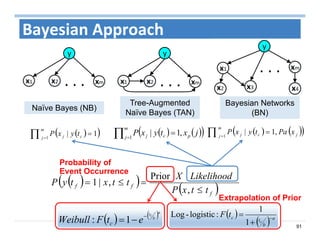
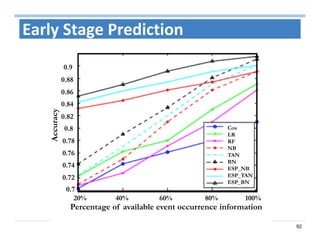
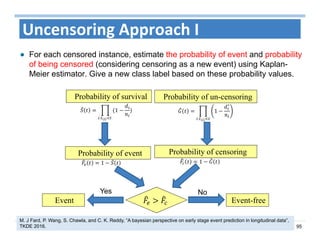
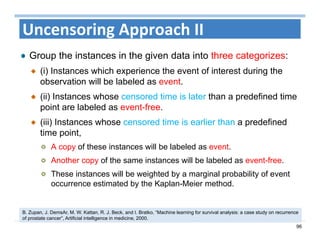
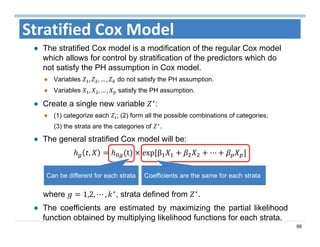
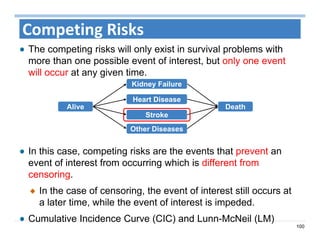
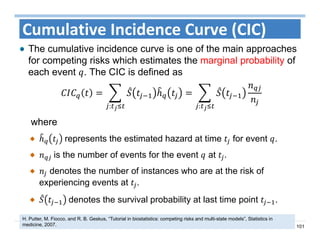
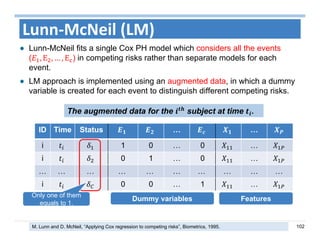
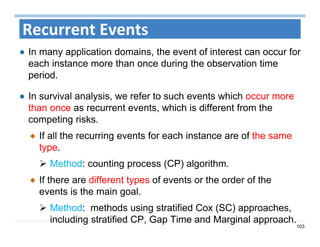
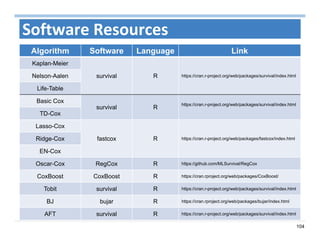
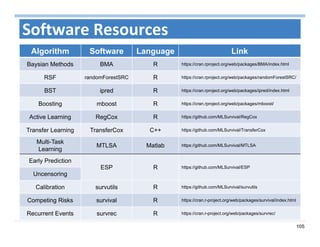
The document outlines the use of machine learning methods for survival analysis, discussing various modeling applications including healthcare event prediction, student dropout, and crowdfunding project success. It covers key statistical methods used in survival analysis, describes evaluation metrics for assessing performance, and highlights the importance of understanding censoring in survival data. Additionally, it provides an overview of various survival analysis methods, including non-parametric, semi-parametric, and parametric approaches.