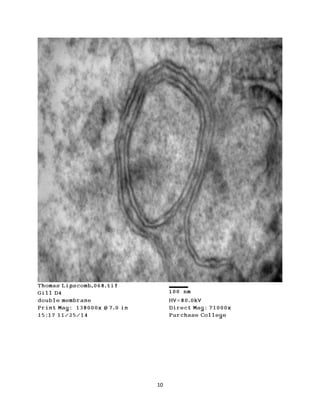
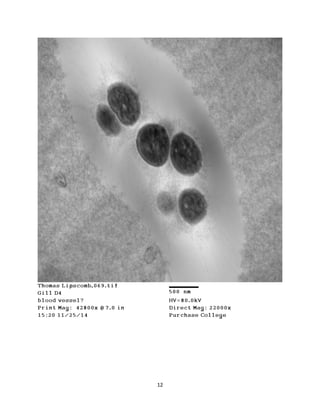
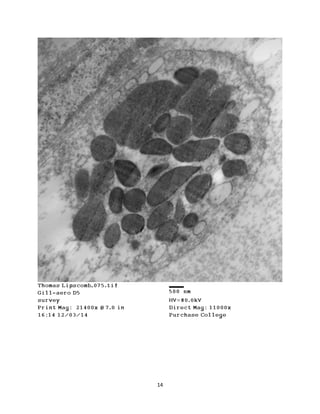
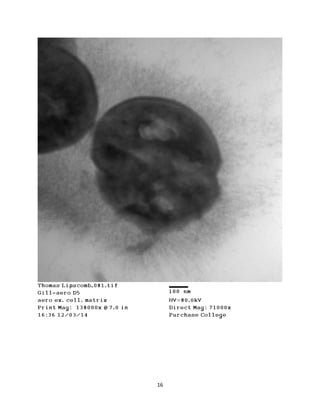
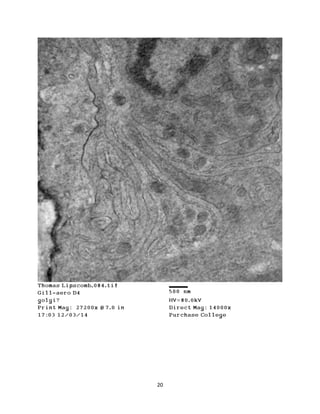
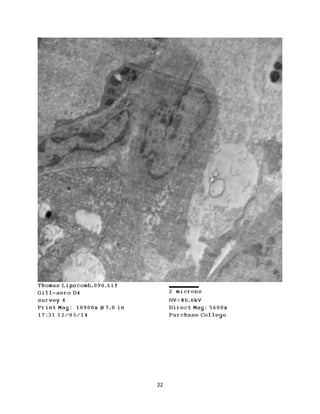
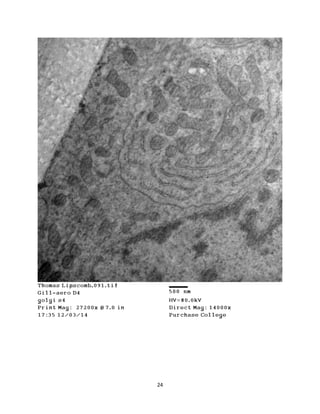
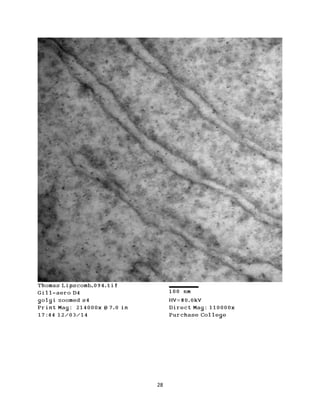
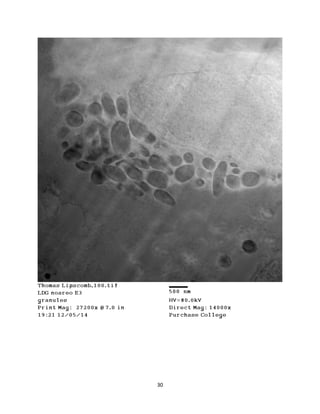
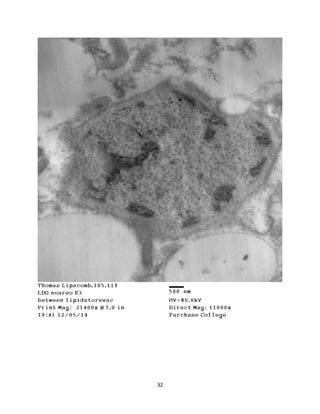
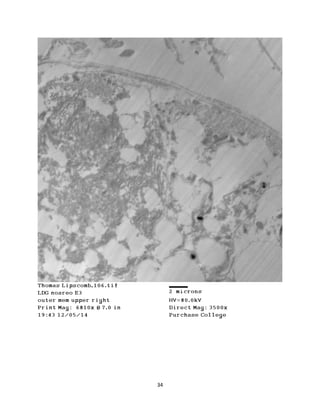
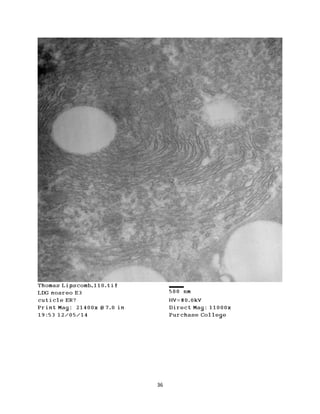
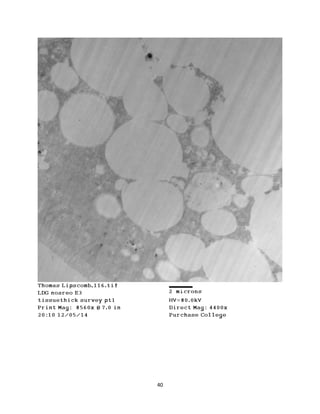
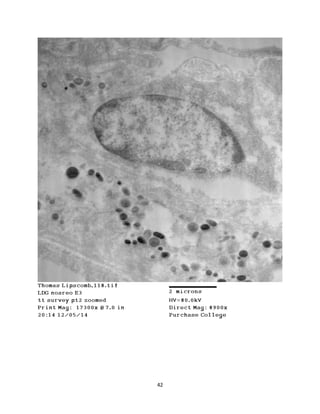
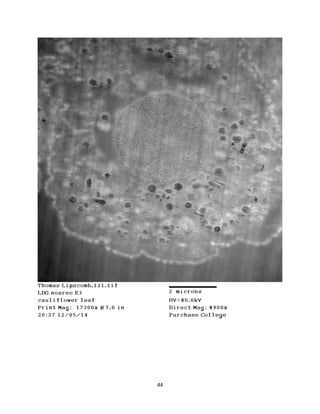
This document summarizes transmission electron microscope (TEM) images of two lobster tissues: the digestive gland and gill infected with Aerococcus viridans bacteria. The author provides background on the lobster anatomy and pathology of A. viridans infection. Methods are described for sample preparation including fixation, dehydration, embedding and sectioning for TEM imaging. Images were taken of the non-infected digestive gland as a control and the infected gill to compare tissue ultrastructure and pathology between the two.