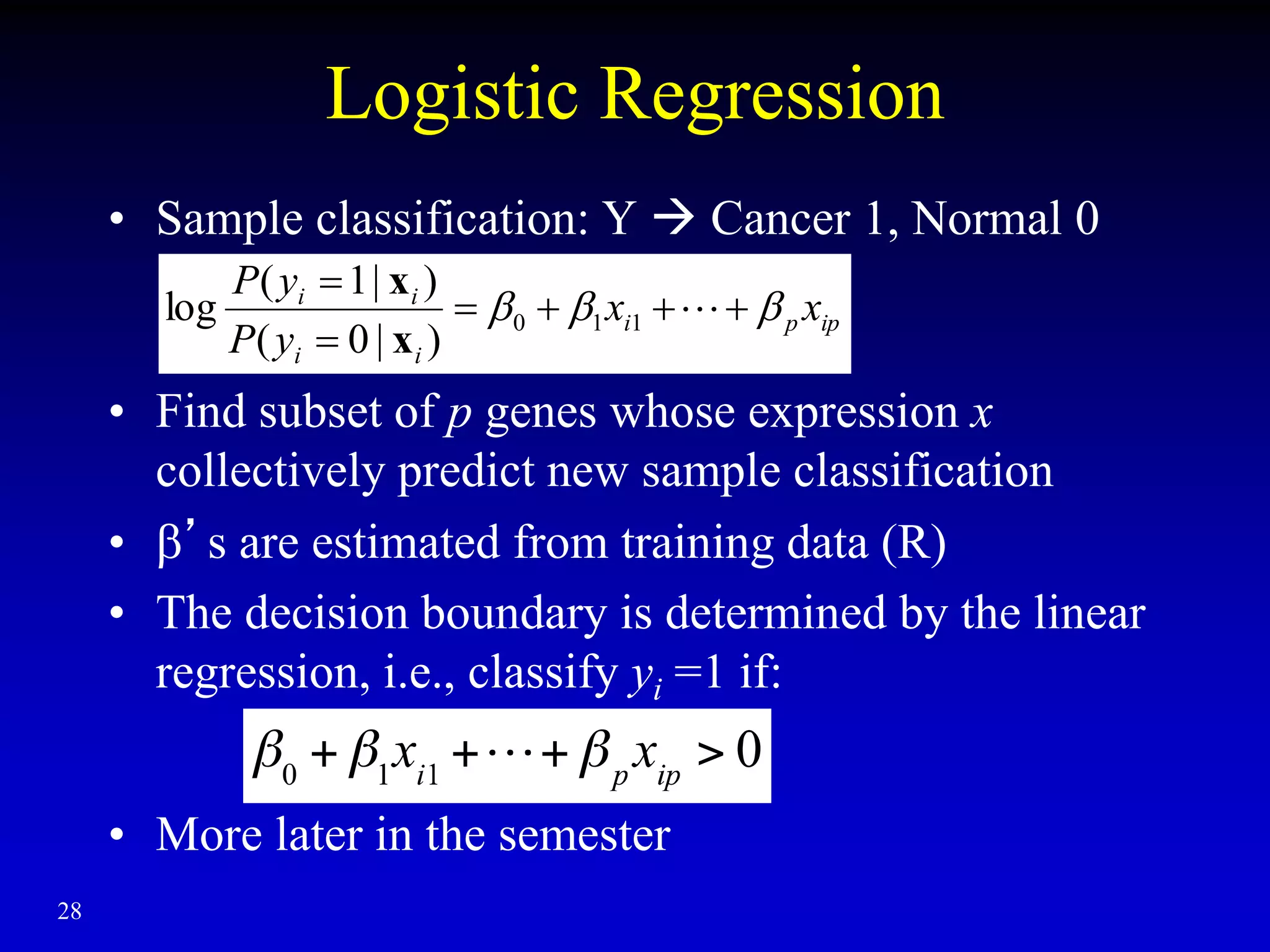
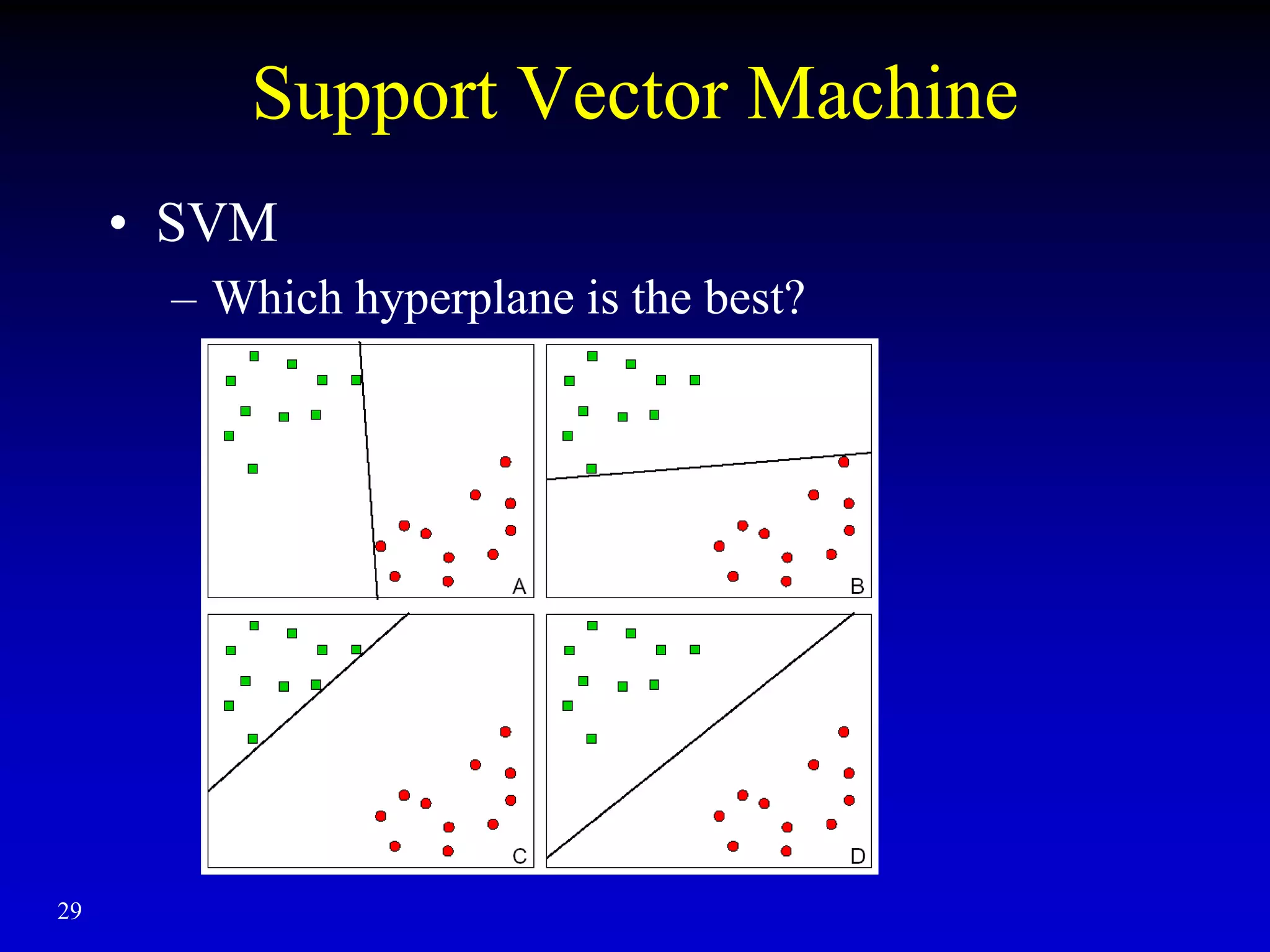
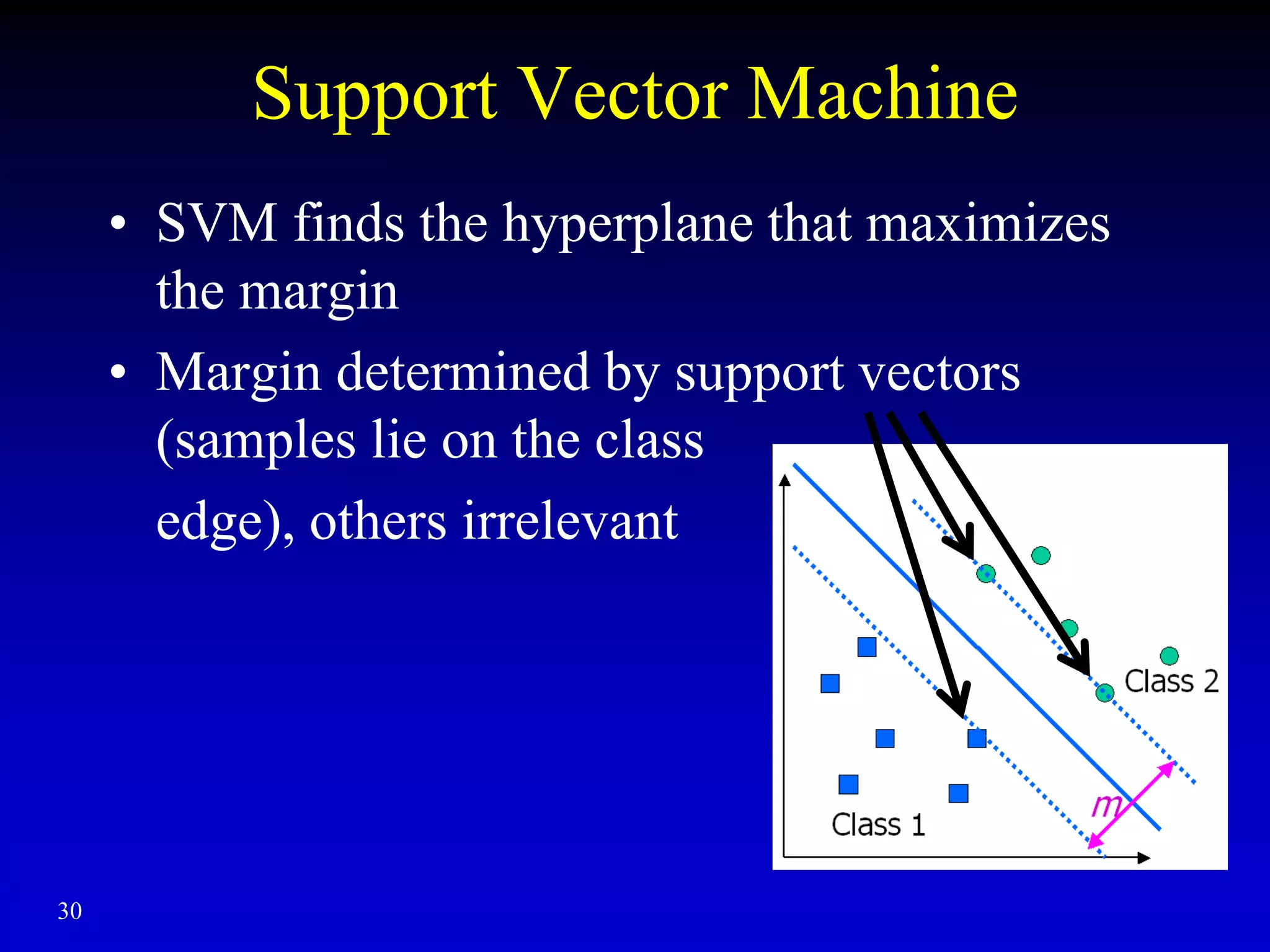
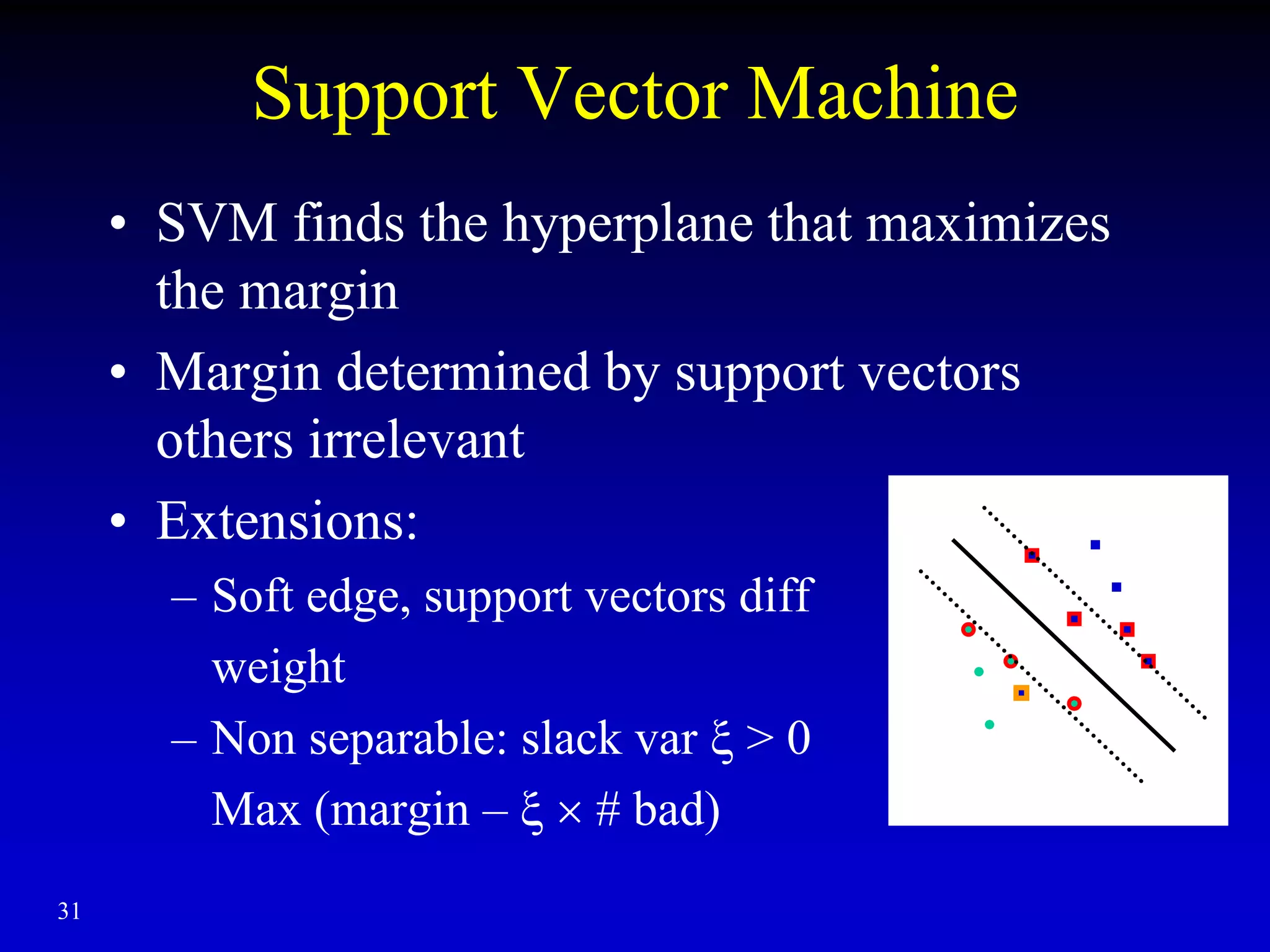
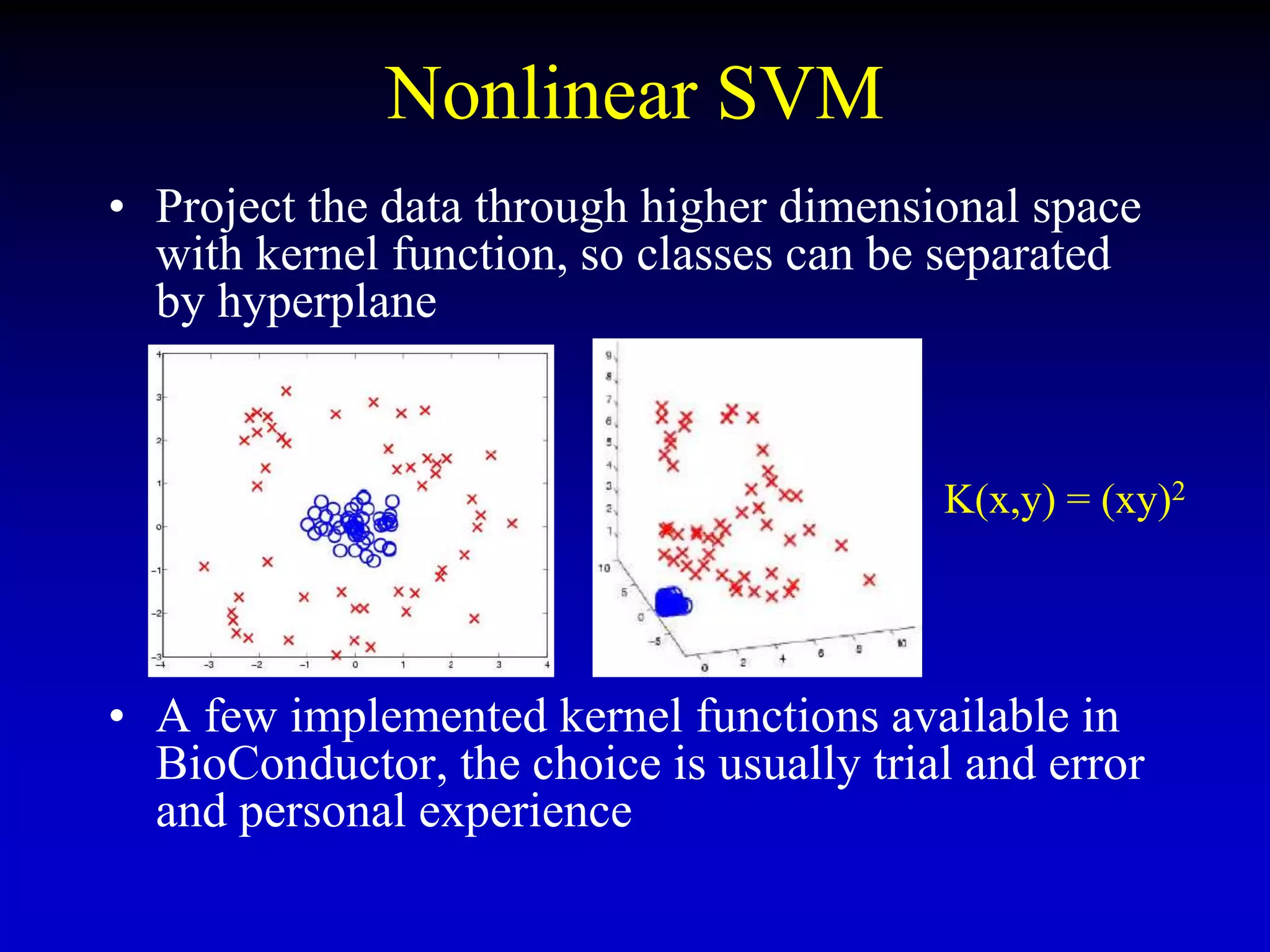
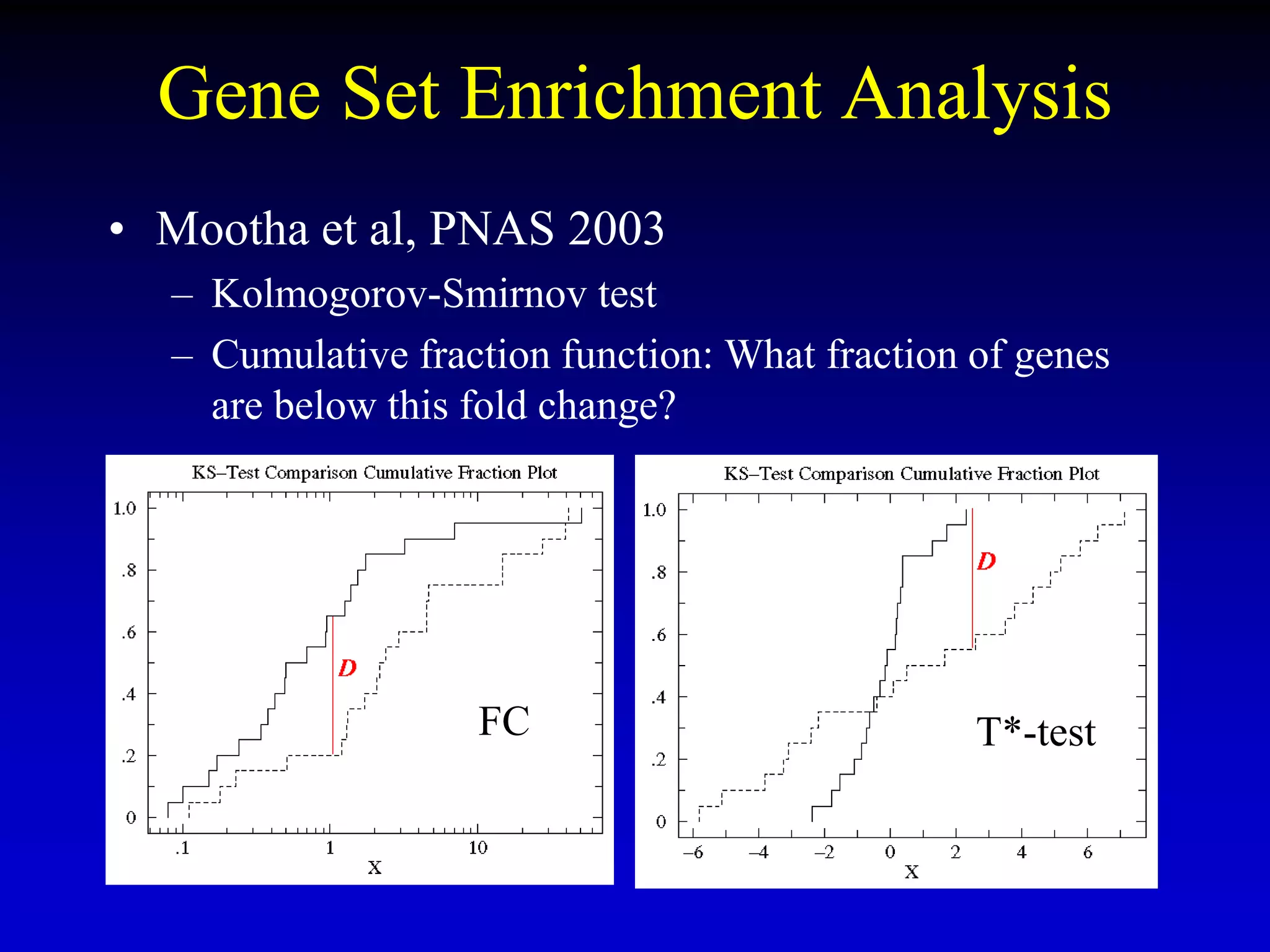
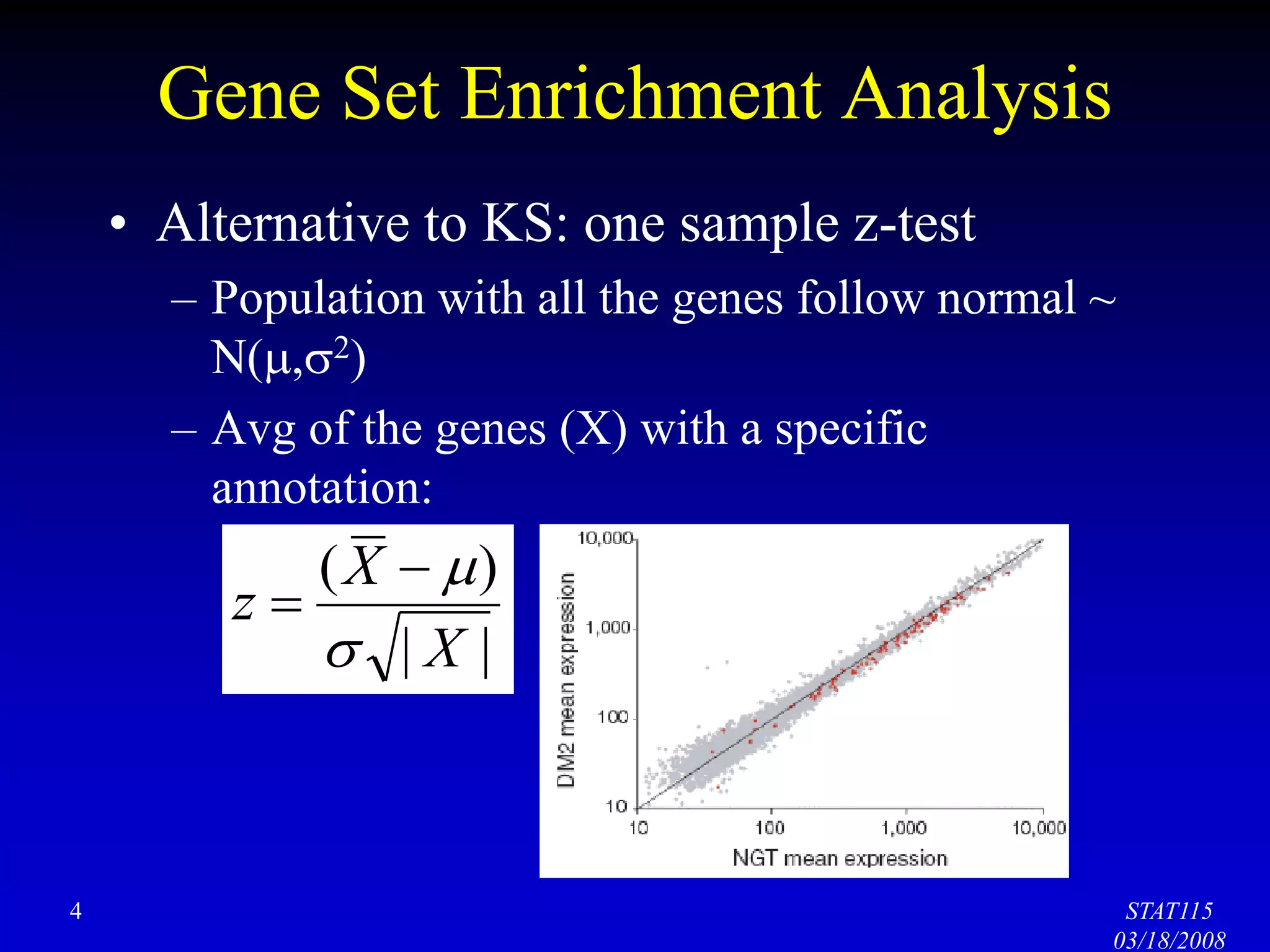
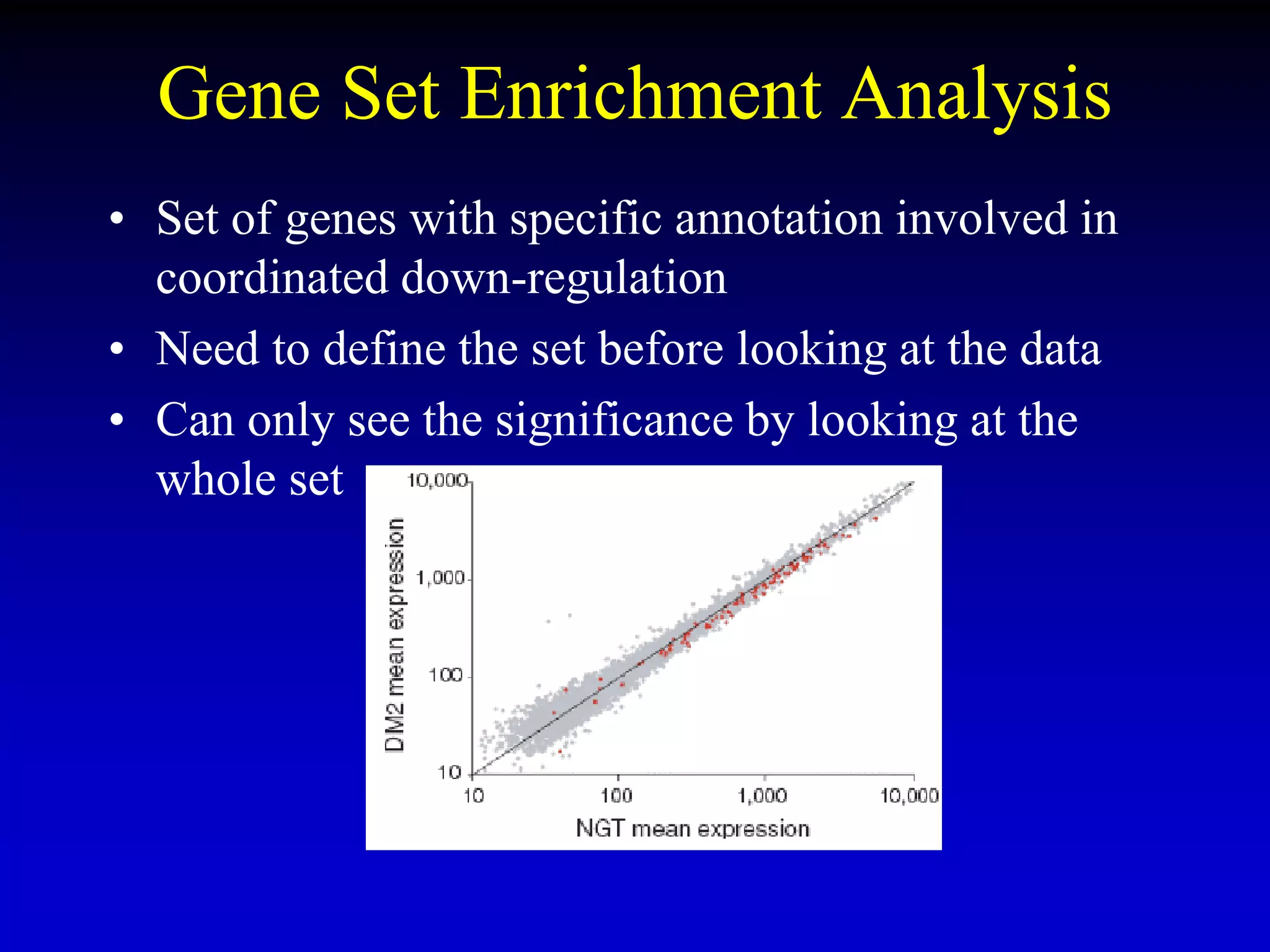
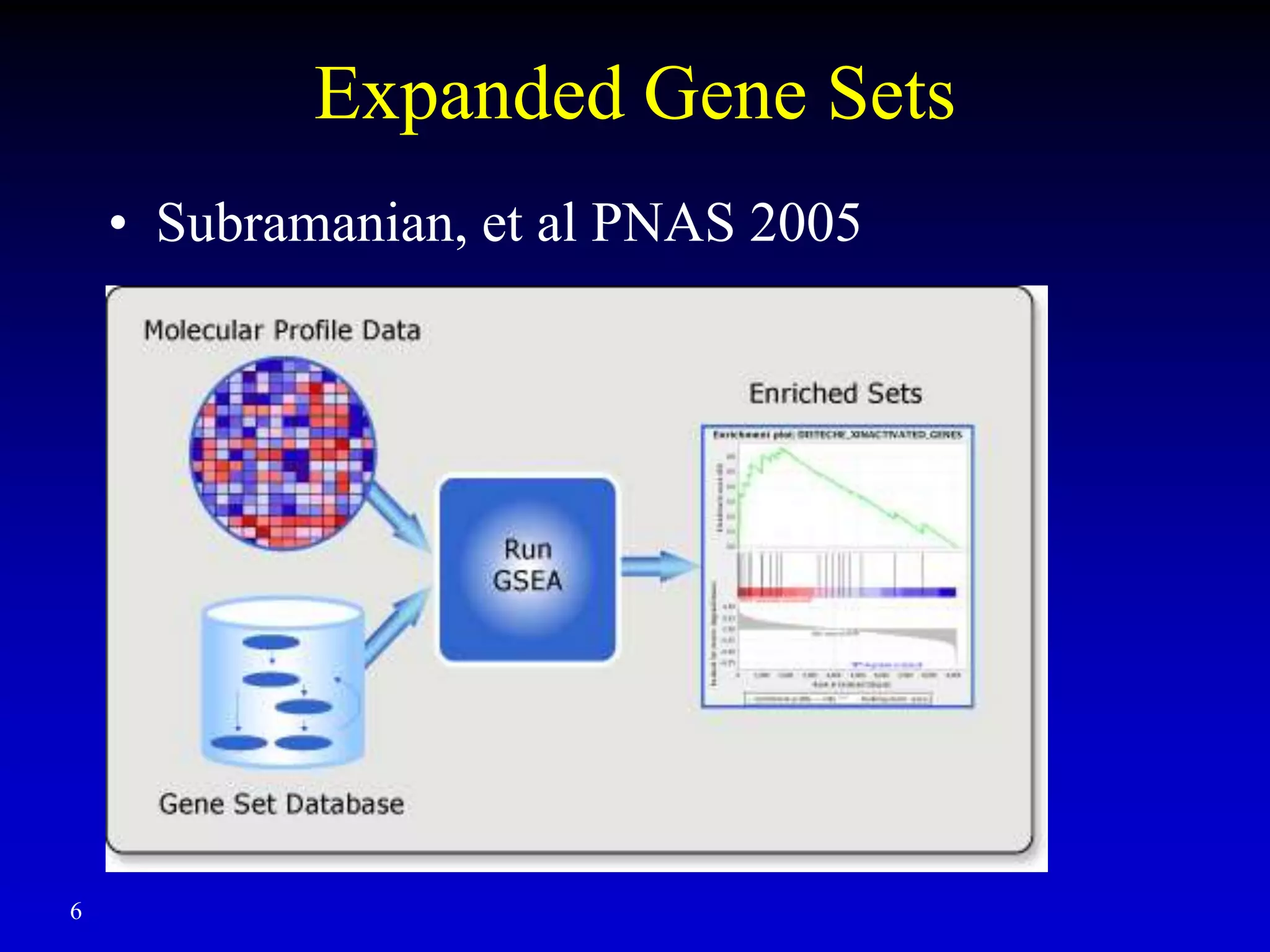
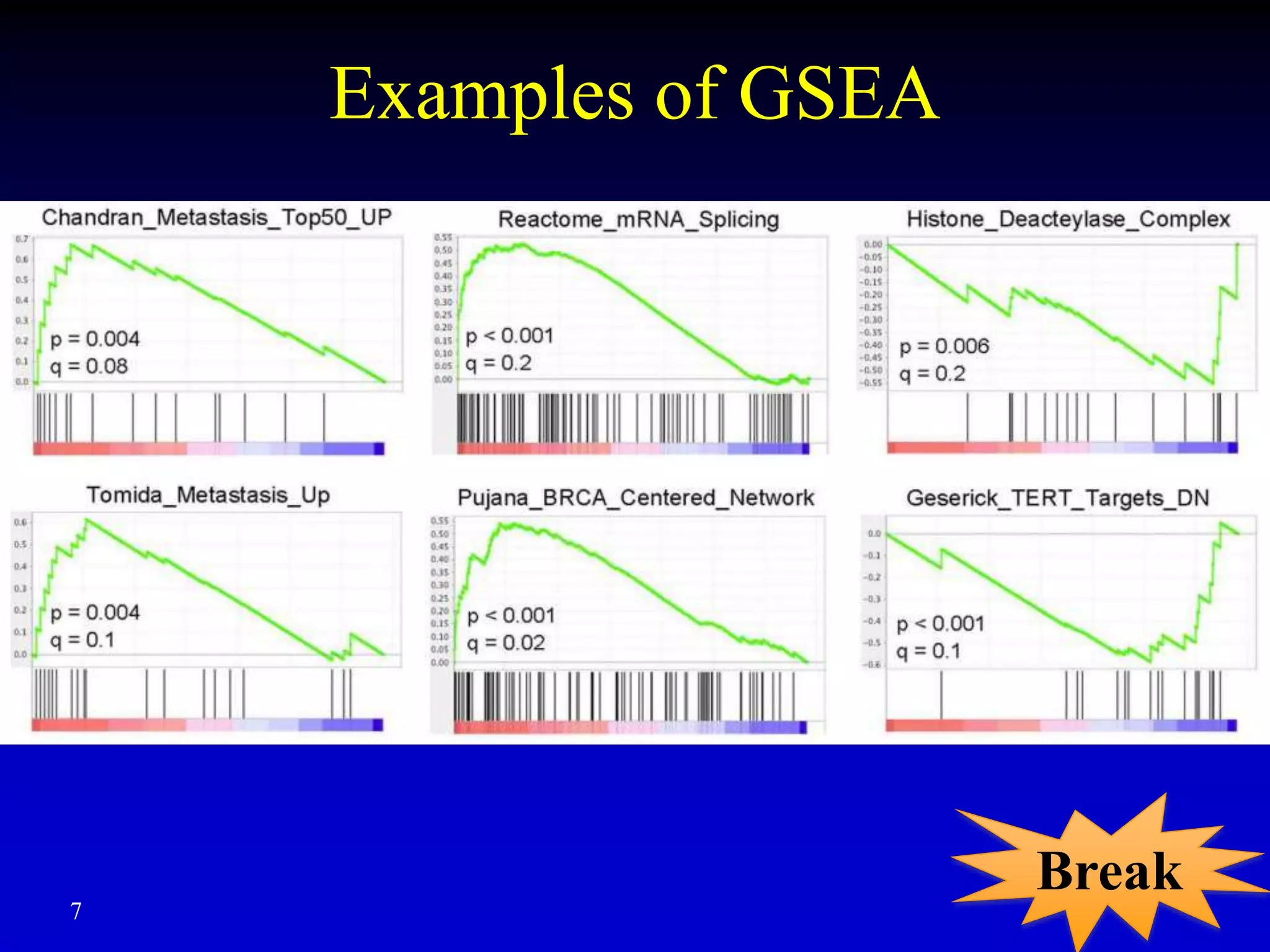
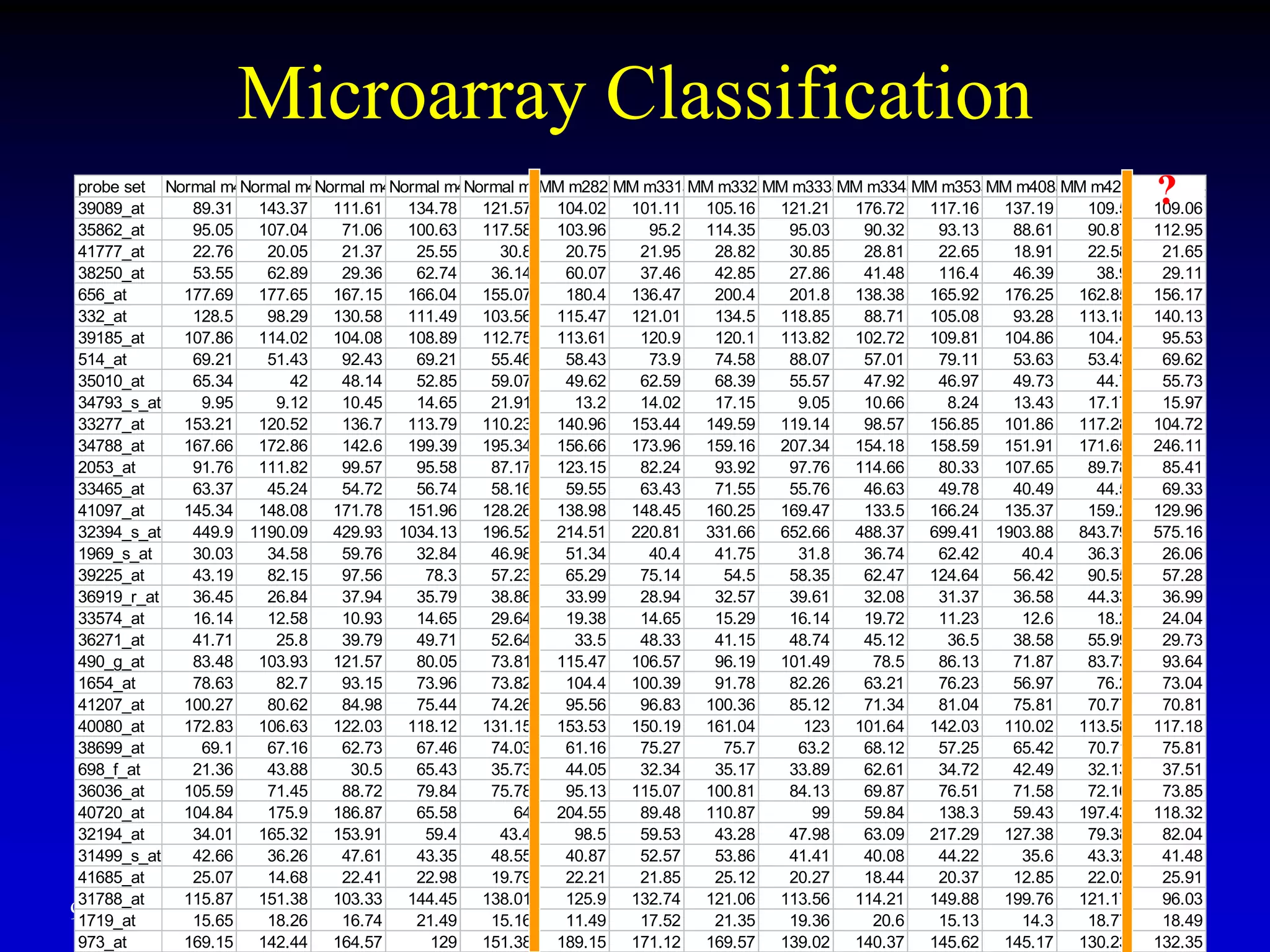
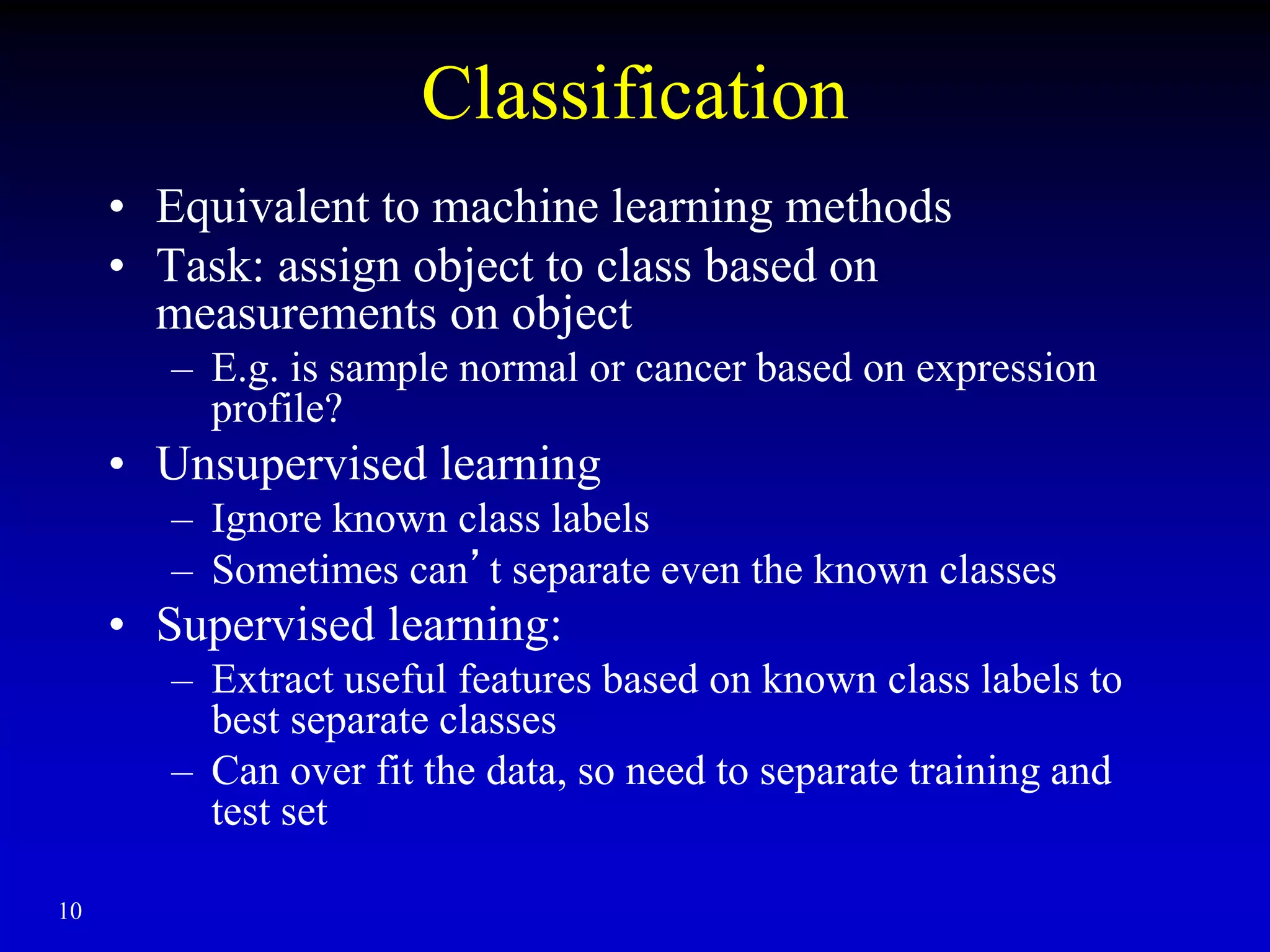
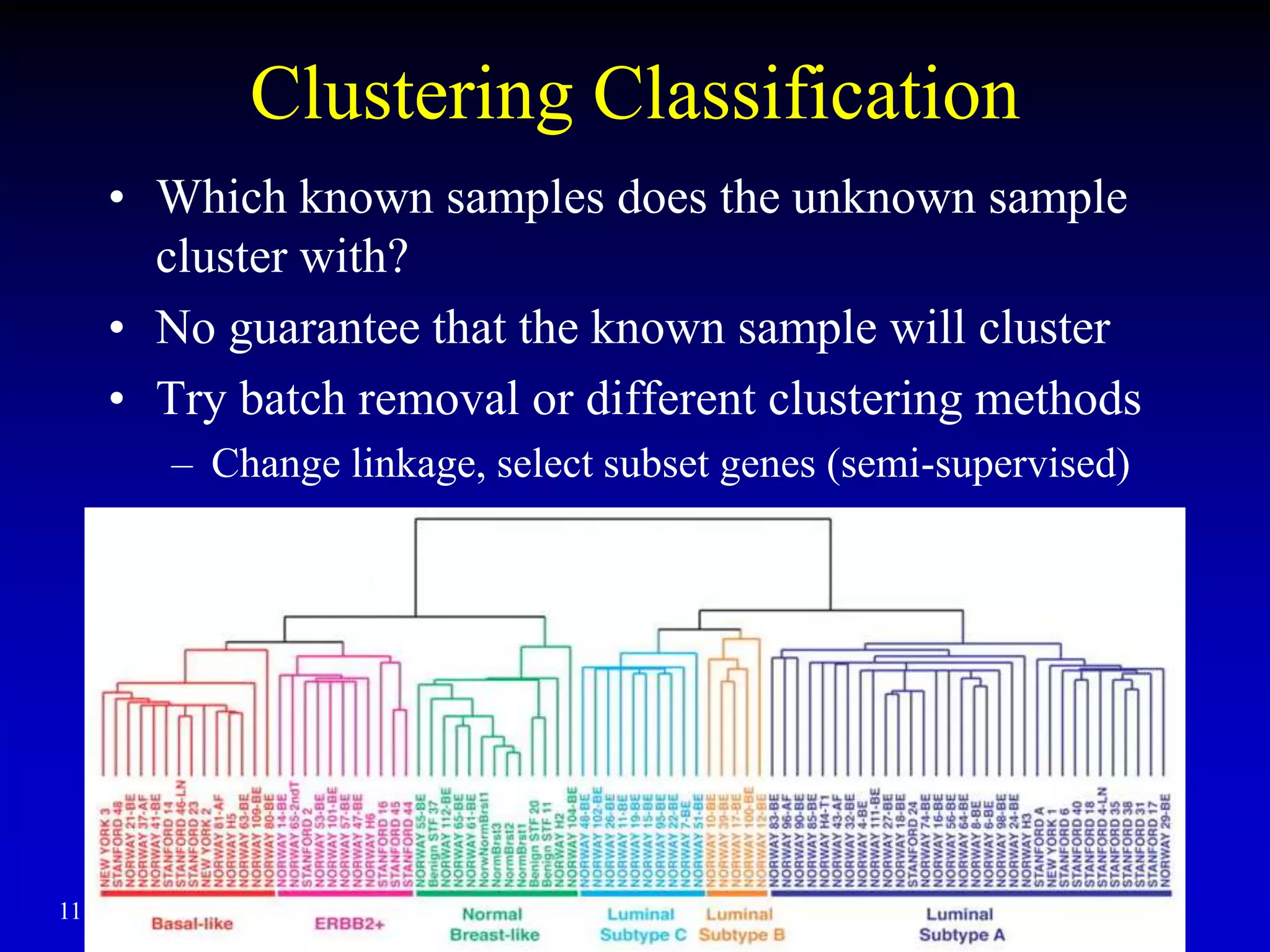
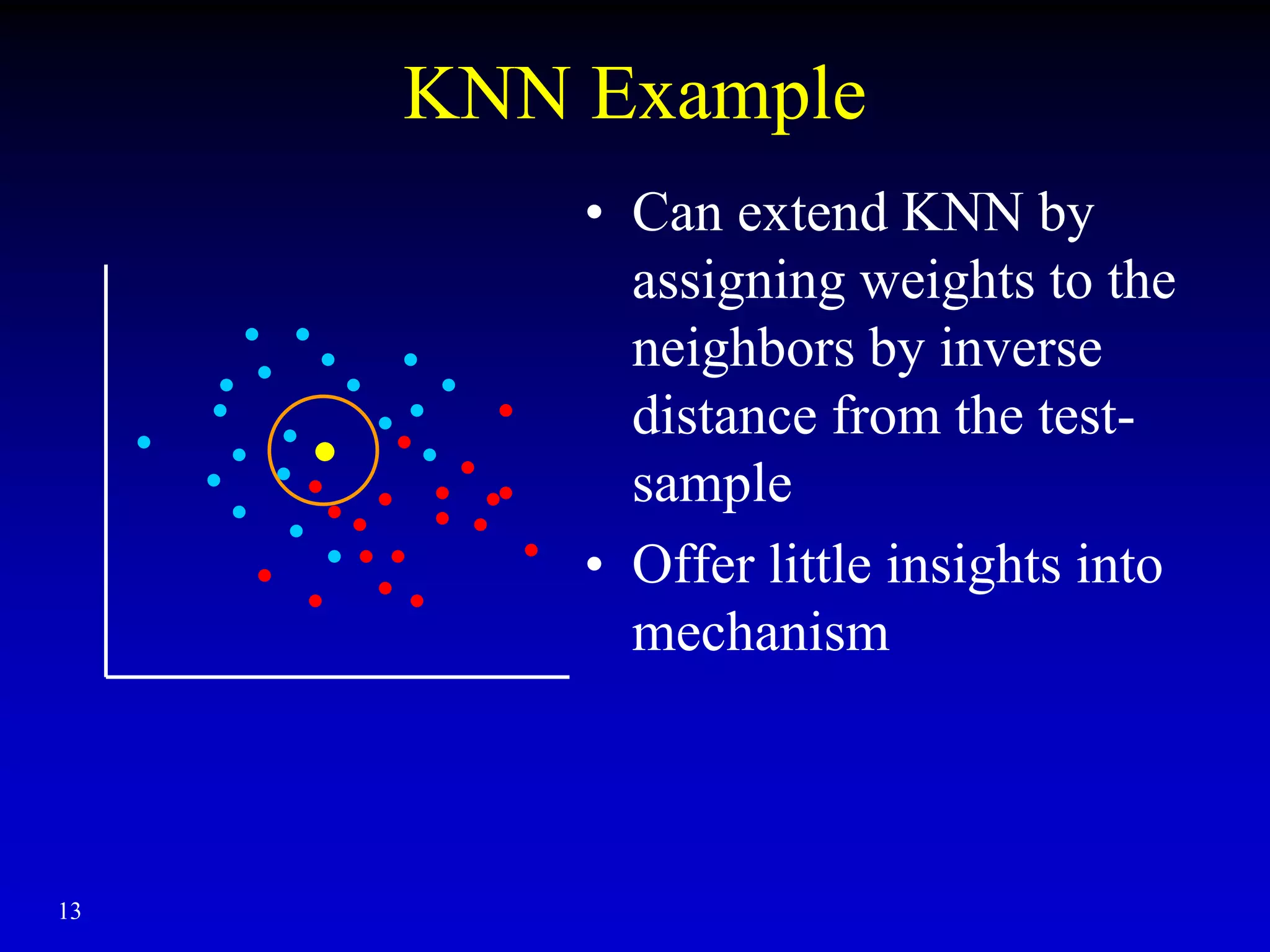
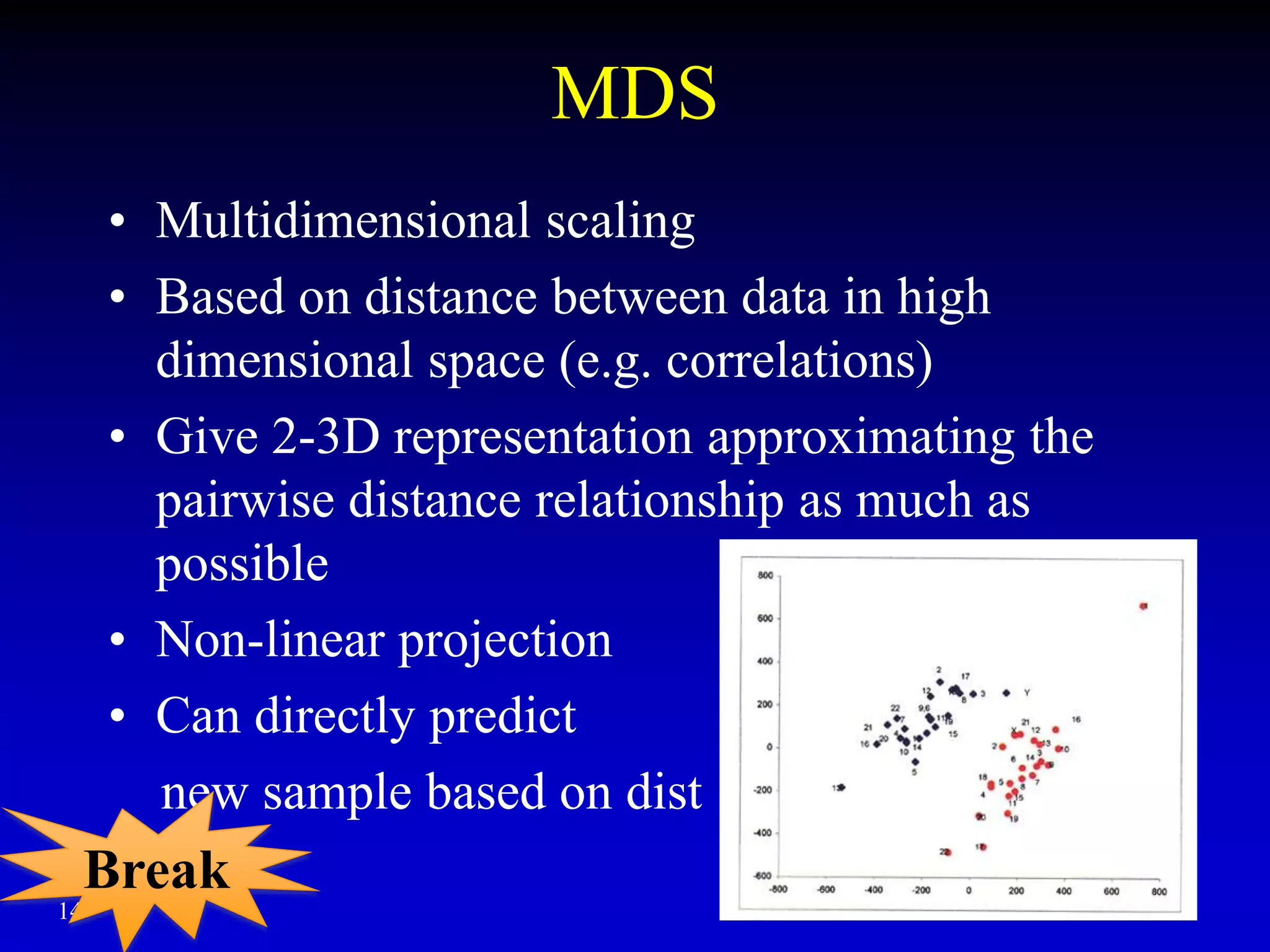
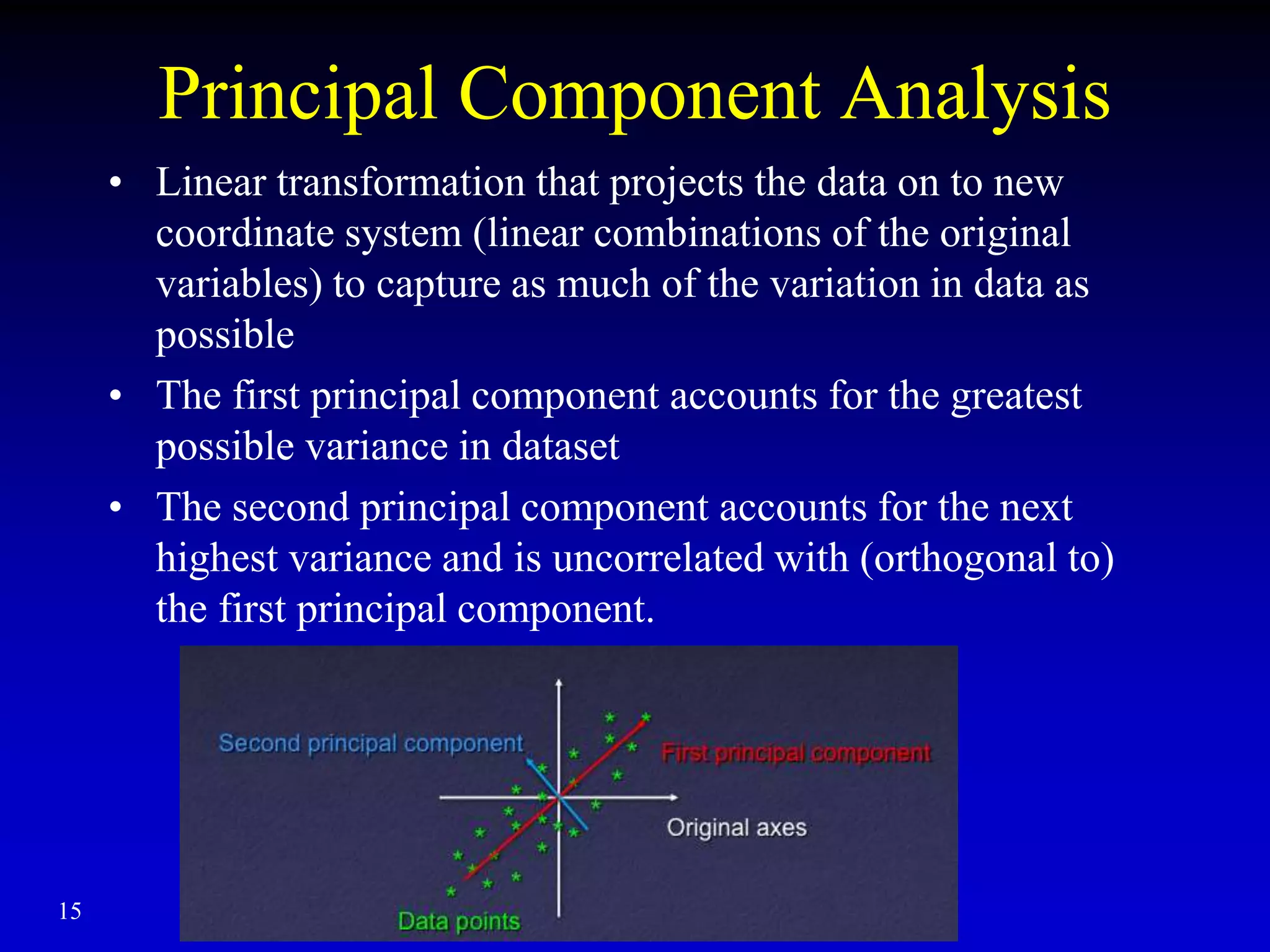
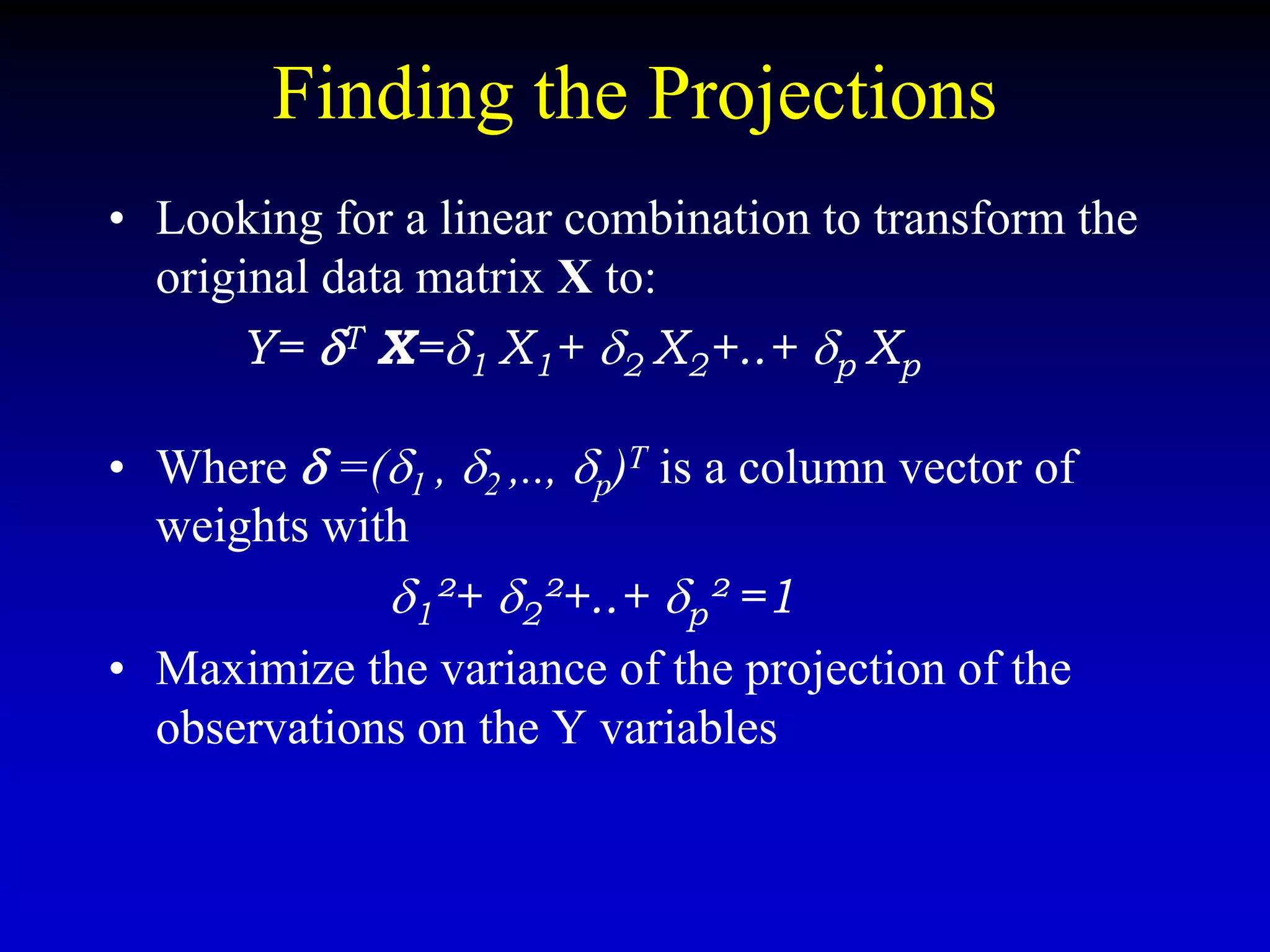
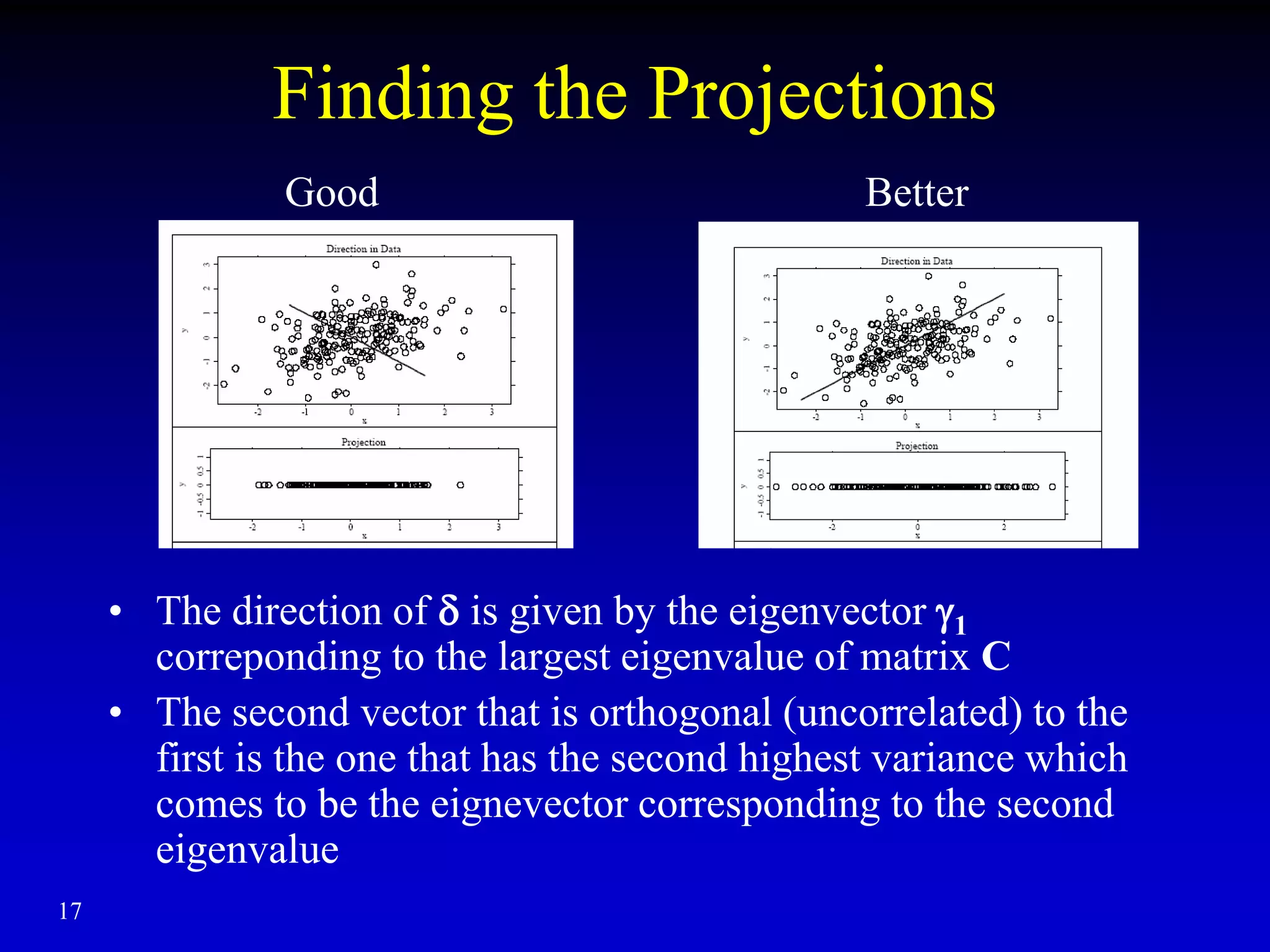
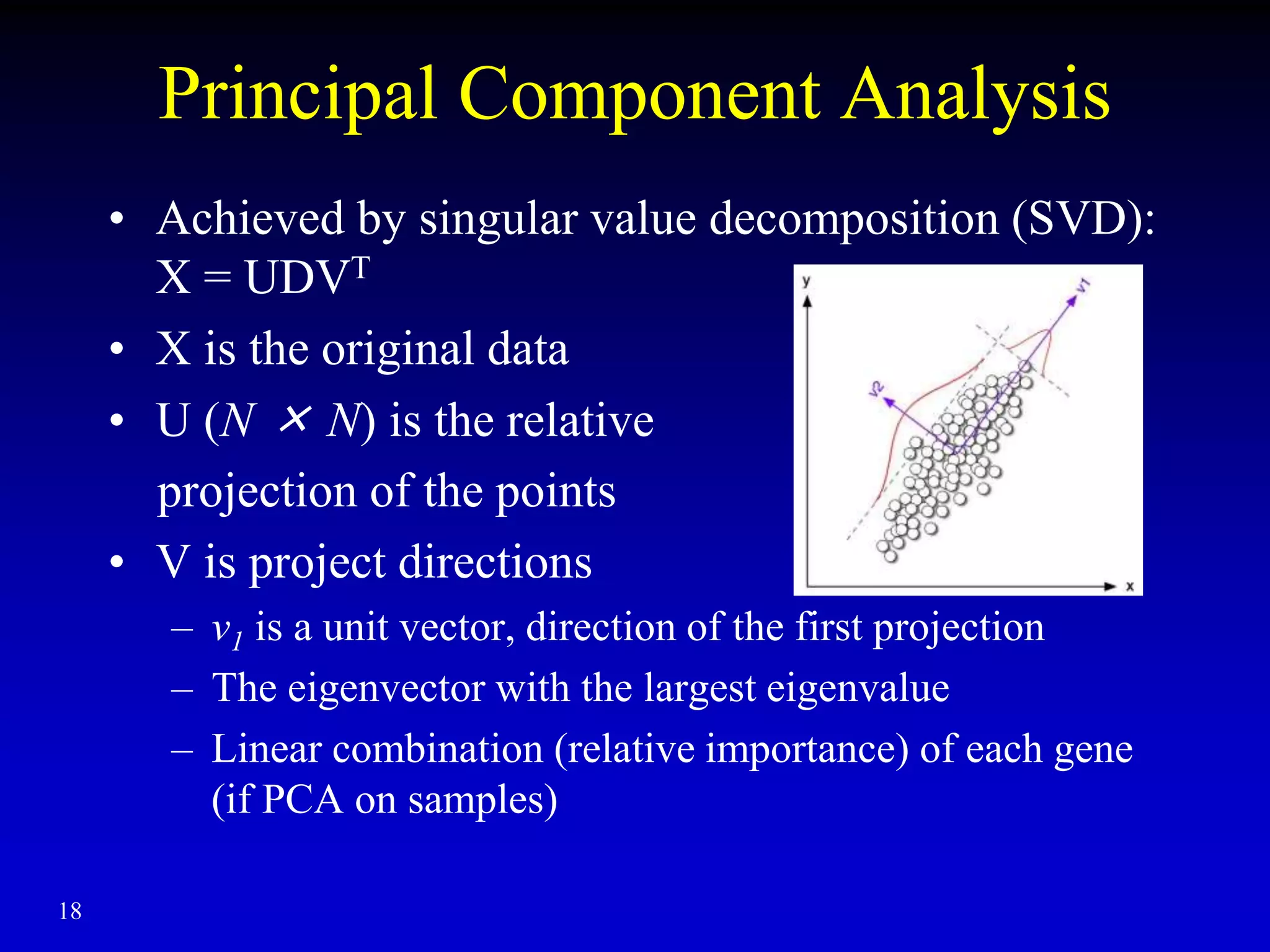
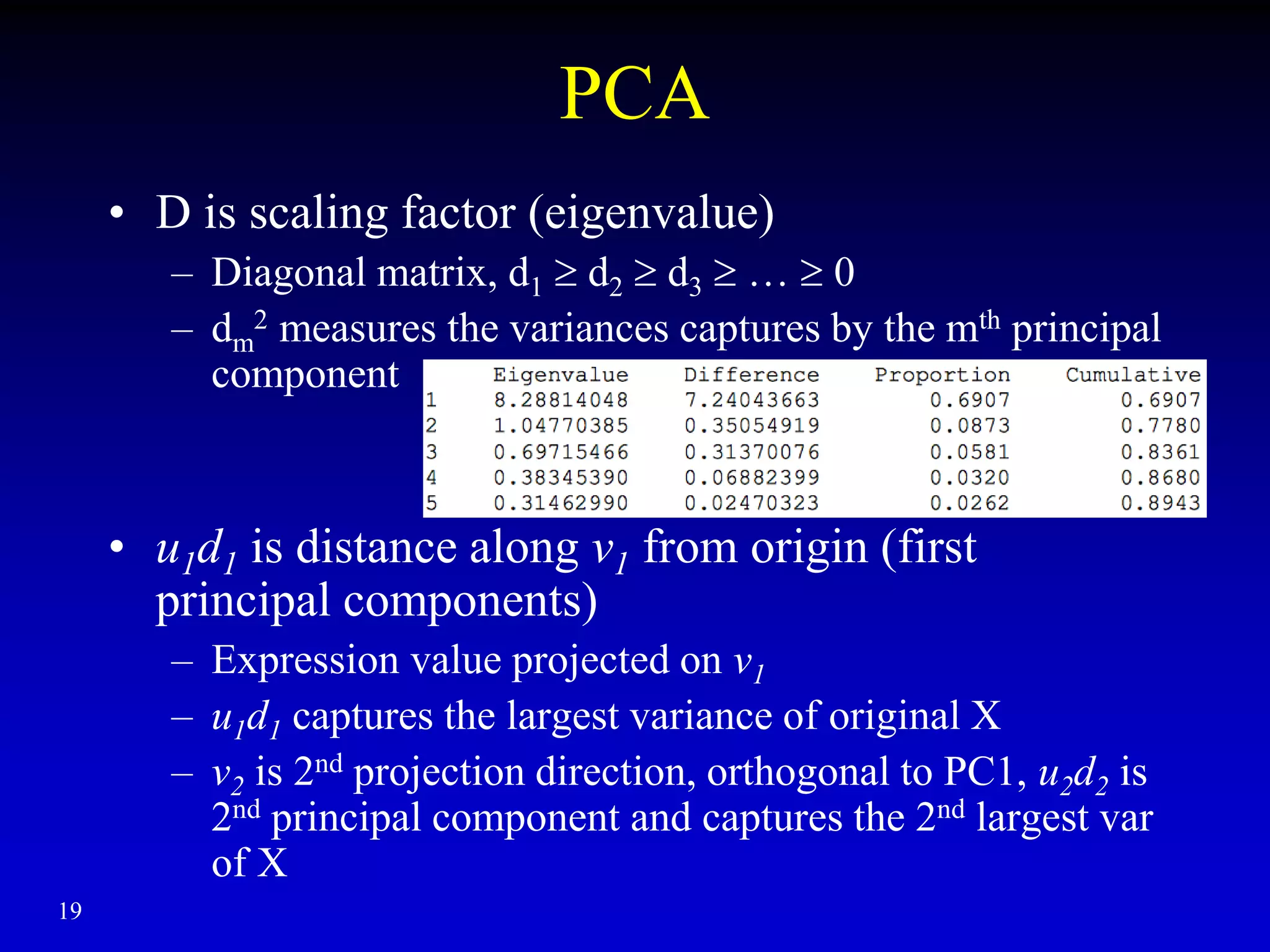
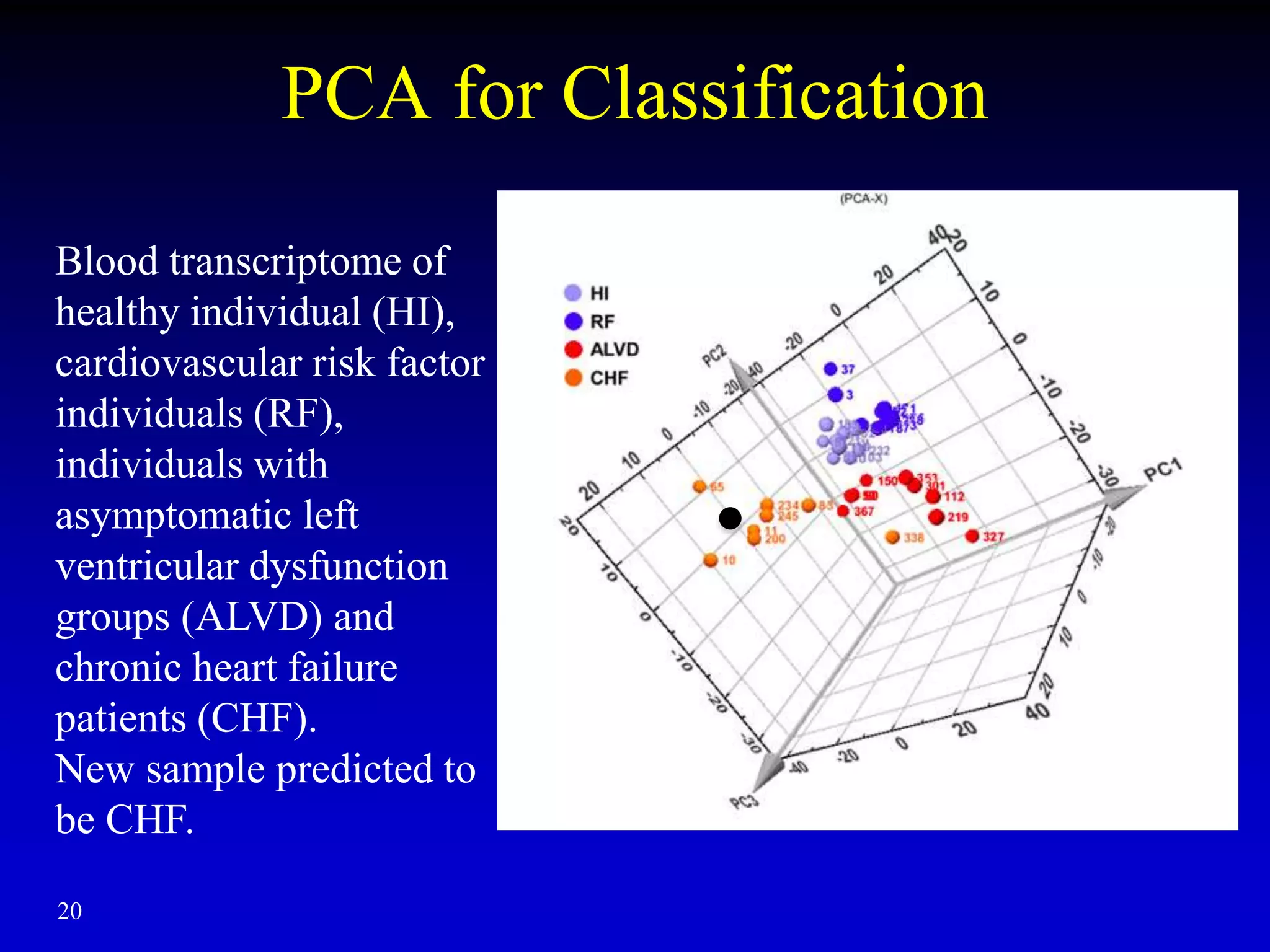
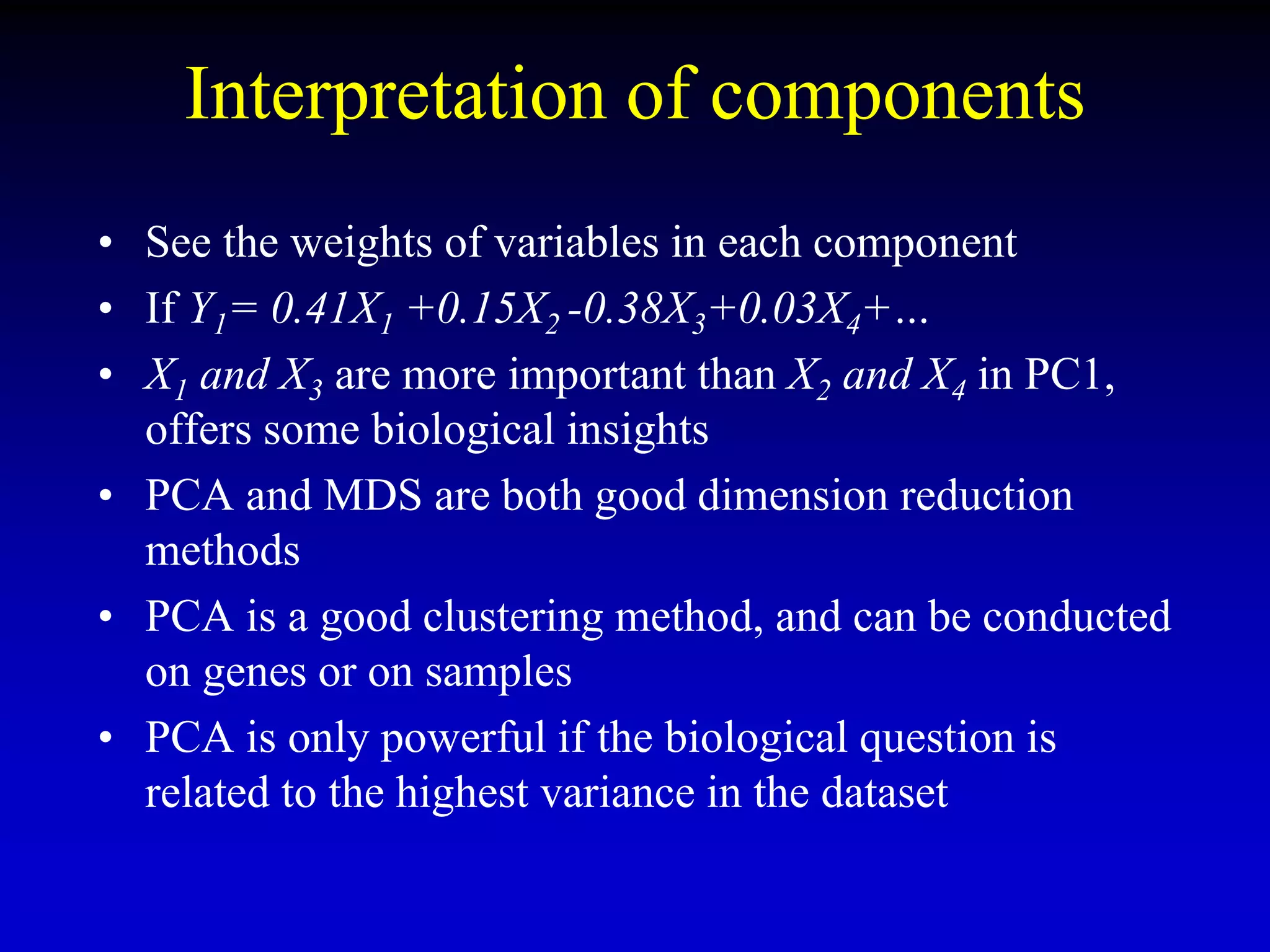
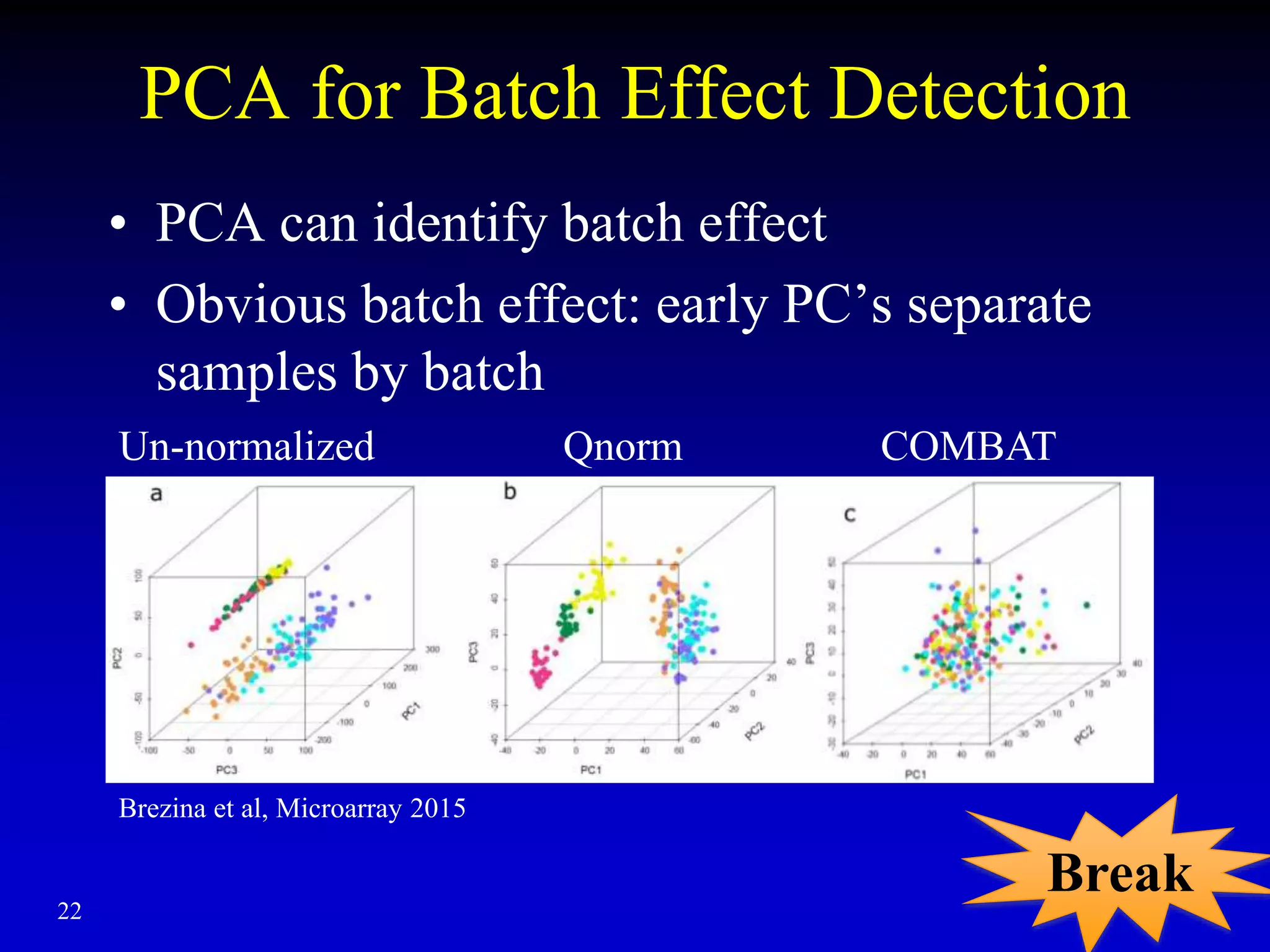
This document discusses gene set enrichment analysis (GSEA). GSEA can be used to analyze microarray data when no single gene is significantly differently expressed between two conditions, but a group or set of genes shows slight coordinated differences. GSEA checks for enrichment of specific gene sets, such as those involved in similar biological processes, by comparing the expression values of genes within the set to all other genes using statistical tests like the Kolmogorov-Smirnov test. The document provides examples of applying GSEA and discusses related methods like expanded gene set analysis.





![Example (wiki)
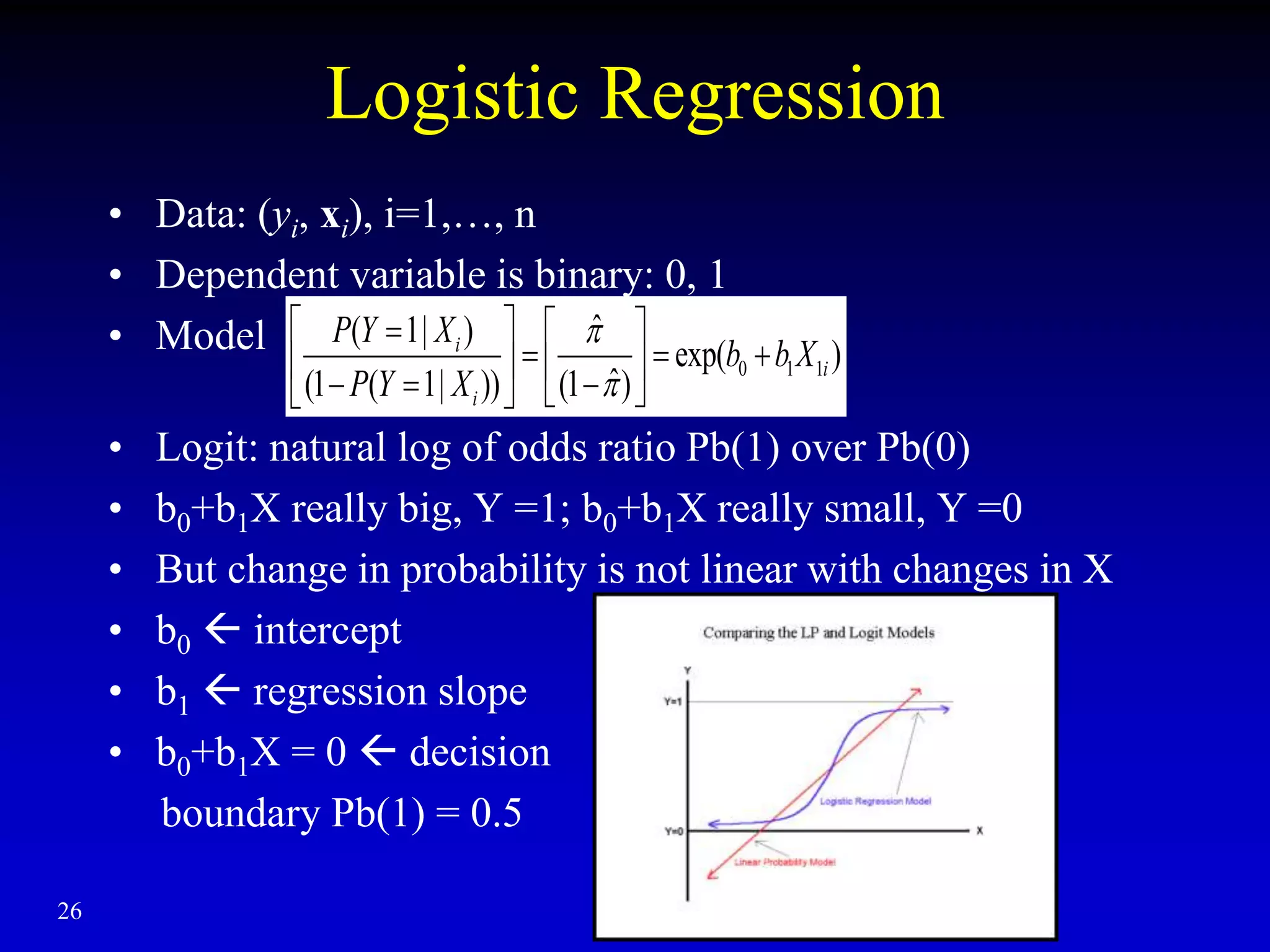
• Hours study on Pb of passing an exam
• Significant association
– P-value 0.0167
• Pb (pass) = 1/[1+exp(-b0-b1X)]
= 1/[1+exp(4.0777-1.5046* Hours)]
• 4.0777/1.5046 = 2.71 hours Pb (pass) = 0.5
27](https://image.slidesharecdn.com/lect5gseaclassify1-220919150739-0cf7bca8/75/Lect5_GSEA_Classify-1-ppt-26-2048.jpg)