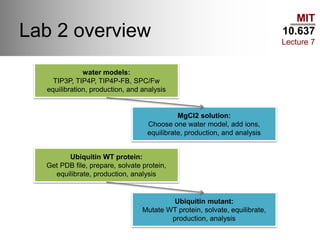
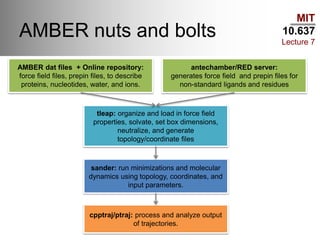
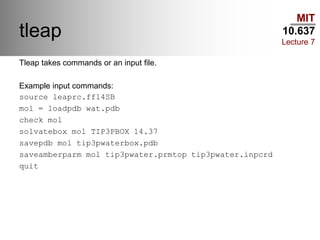
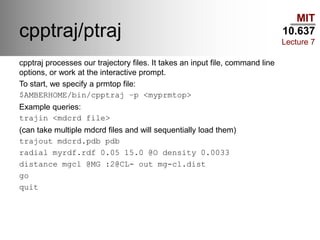
This document outlines the procedures and tools used in a classical molecular dynamics lab as part of MIT's course 10.637. It covers various water models, procedures for preparing and analyzing protein structures, commands for using the Slurm queue on the Maverick machine, and command-line tips for efficient navigation and file management. The content is geared towards helping students set up their computational environment and manage simulations effectively.