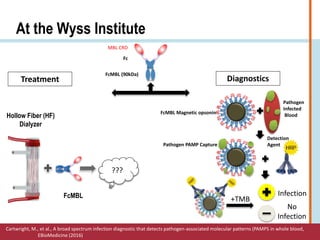
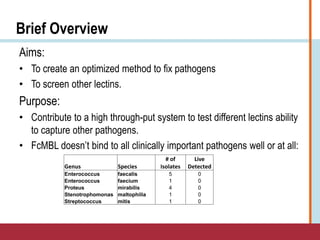
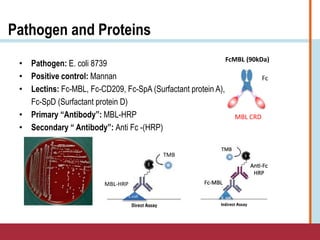
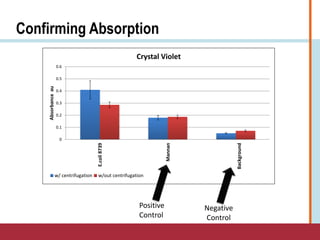
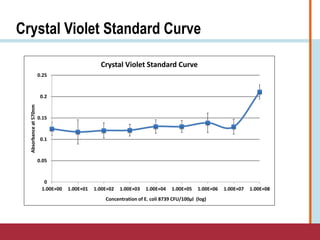
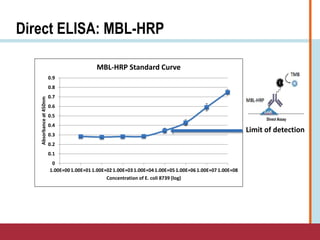
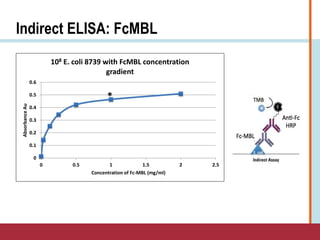
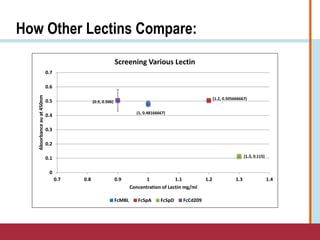
This document summarizes an optimization project to screen lectins for their ability to capture pathogens. The author developed an optimized fixation protocol for pathogens and tested it using crystal violet staining and ELISA assays. ELISA results showed the protocol worked and FcMBL detected E. coli within detectable limits. Other lectins were screened and compared to FcMBL using ELISA. While results were promising, more optimization is needed regarding blocking agents and lectin concentrations. The author acknowledges contributions from mentors and colleagues.