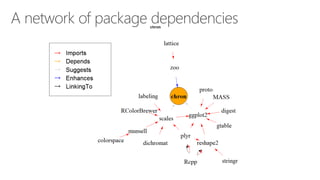
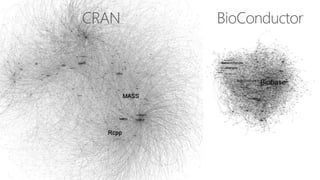
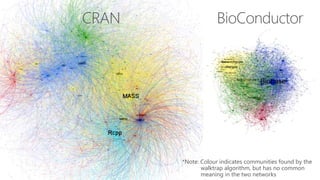
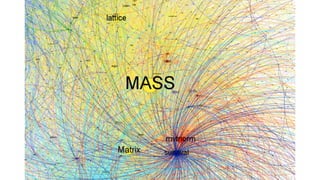
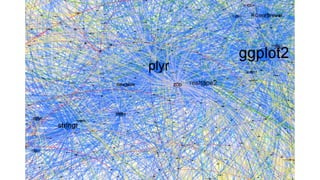
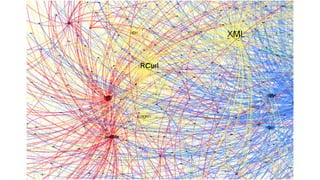
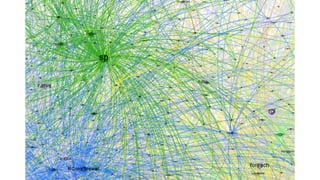
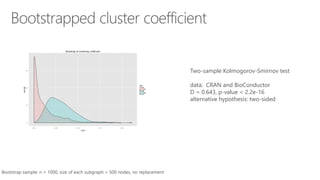
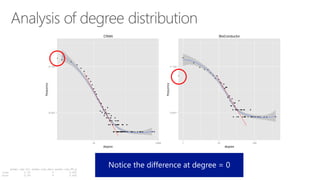
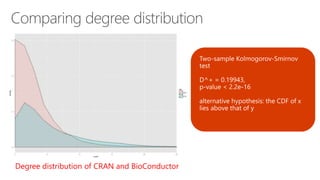
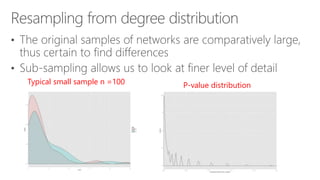
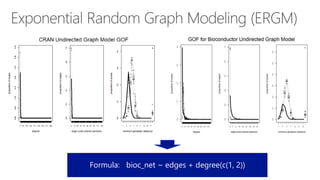
The document compares two biological networks, CRAN and BioConductor, in terms of their topological properties. It finds that CRAN has more nodes and edges than BioConductor, but BioConductor is more densely connected. A statistical test shows the networks have significantly different degree distributions, though both approximately follow power laws.