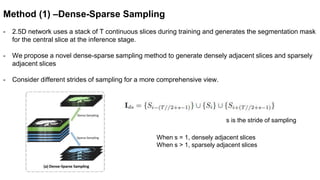
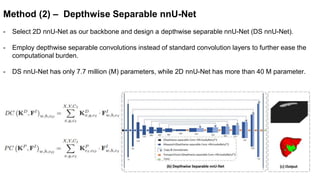
This document presents a novel deep learning framework for efficient liver and tumor segmentation using a multi-slice dense-sparse approach. It highlights the challenges of traditional 2D and 3D networks and introduces a lightweight depthwise separable NNU-Net that offers a significant reduction in parameters while improving accuracy. Extensive experiments validate the effectiveness of the proposed method in both segmentation accuracy and processing efficiency.