1) DNA repair mechanisms are responsible for minimizing damage to DNA from various sources. When DNA damage surpasses a threshold, cells can enter senescence, apoptosis, or become cancerous.
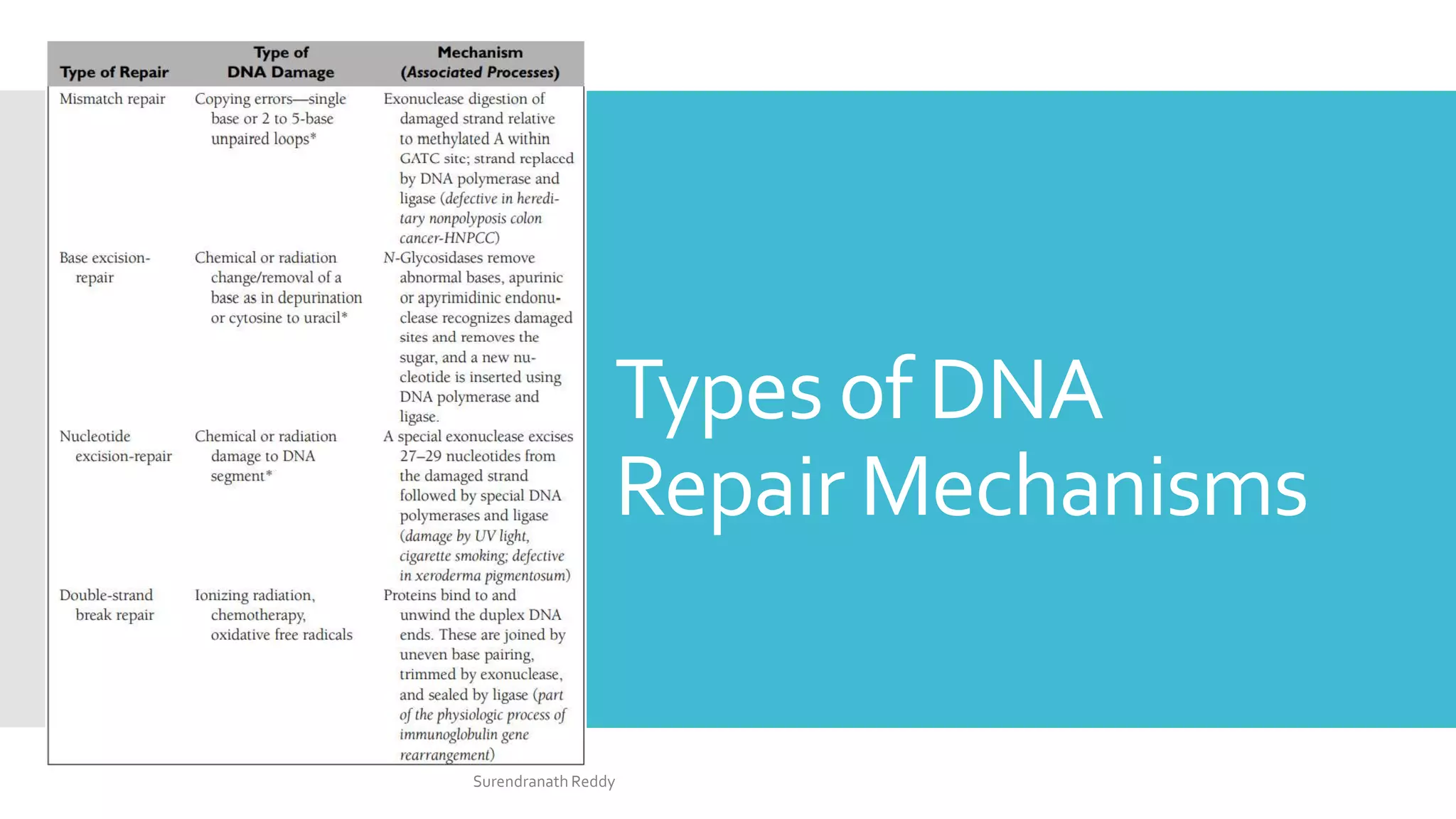
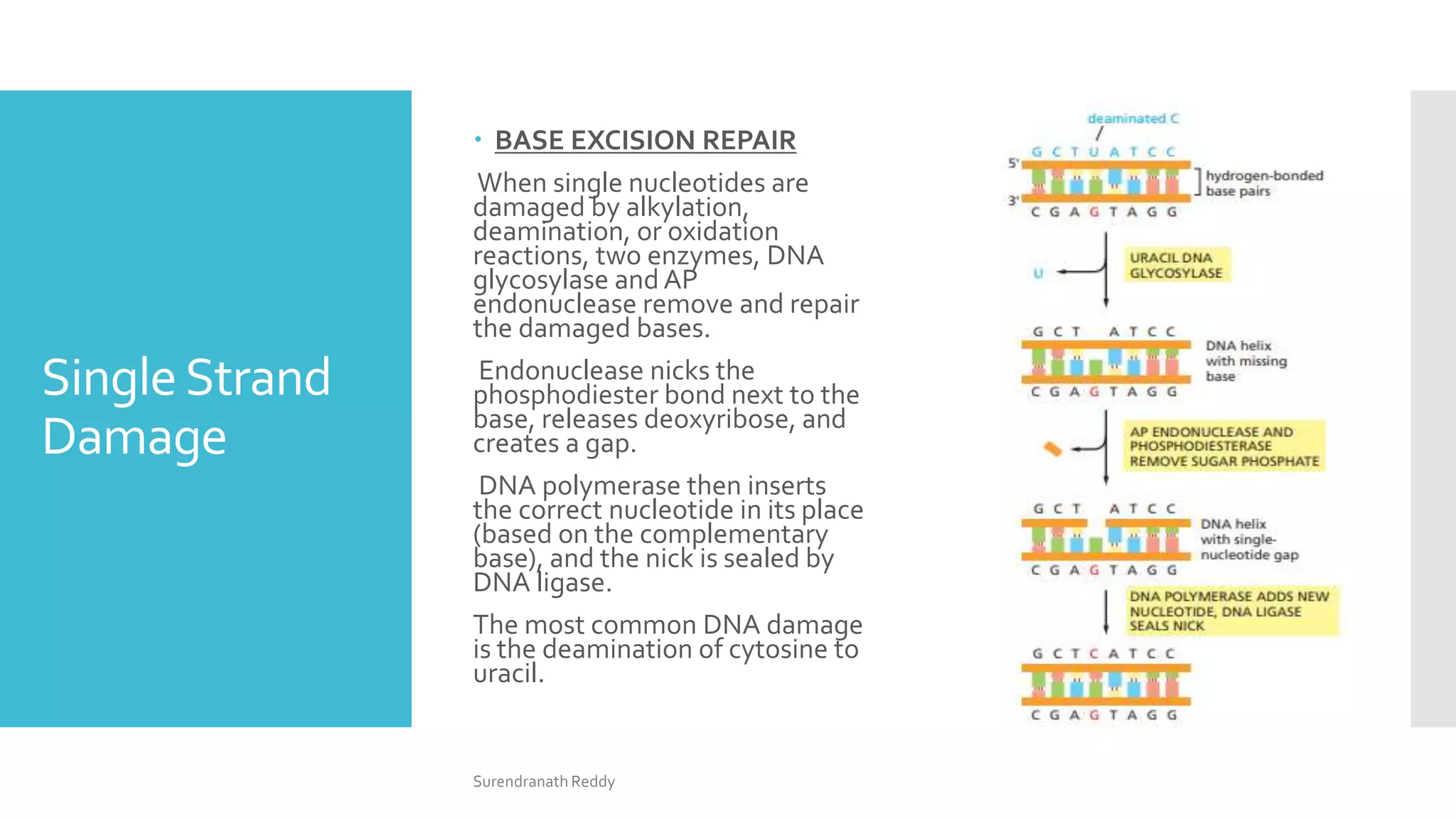
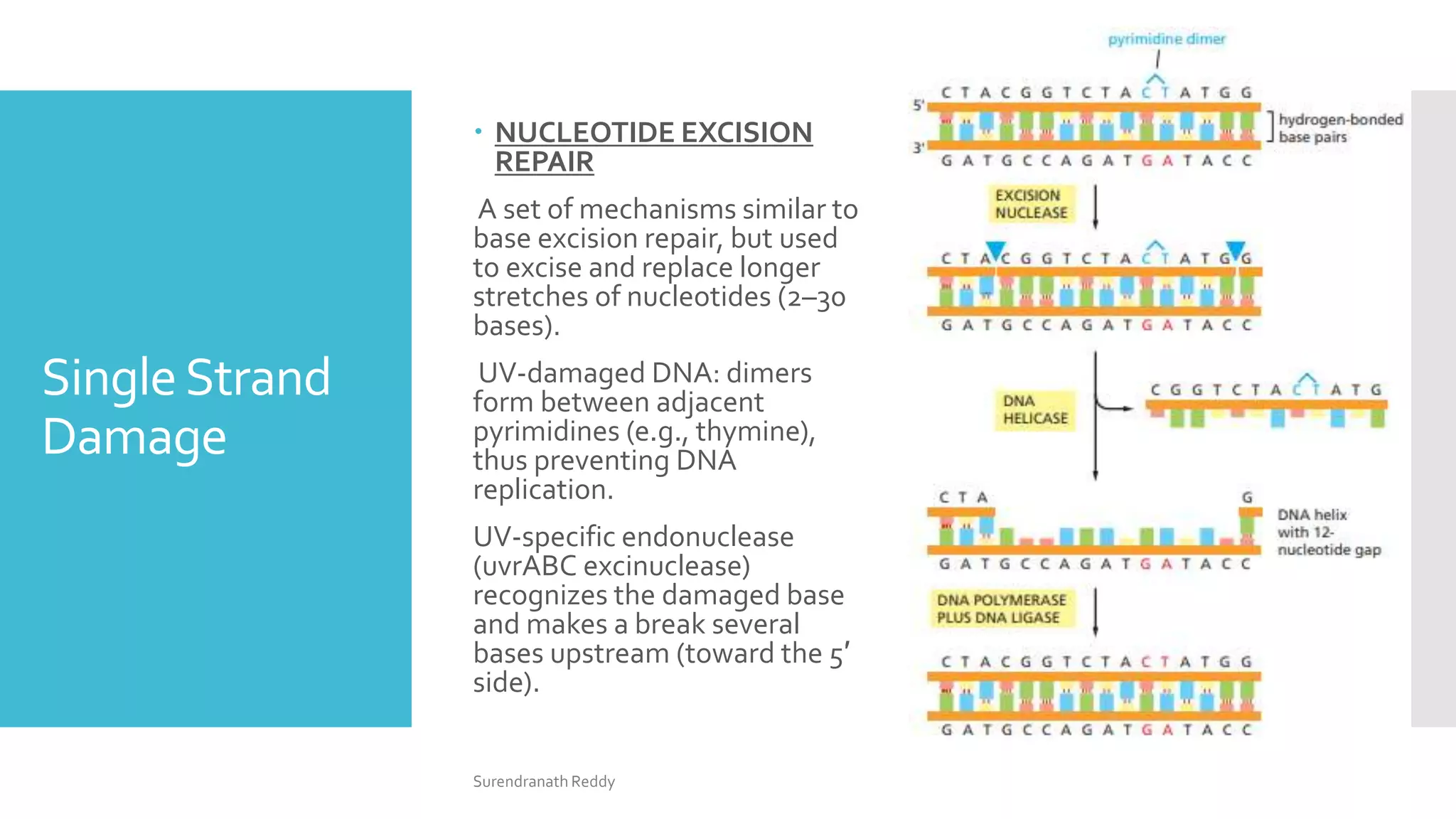
2) There are several DNA repair mechanisms, including base excision repair, nucleotide excision repair, and mismatch repair for single-strand damage, as well as nonhomologous and homologous end joining for double-strand breaks.
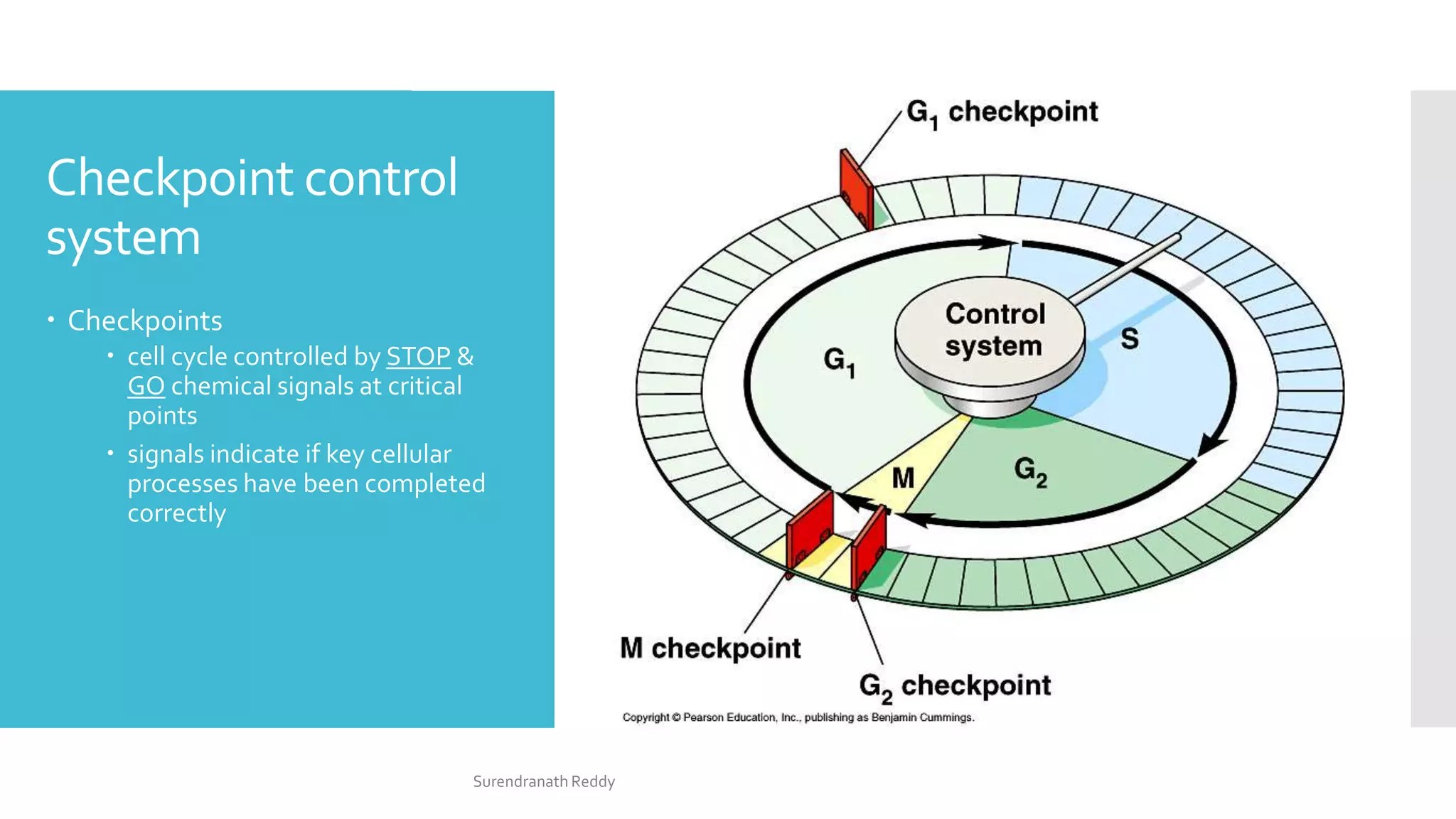
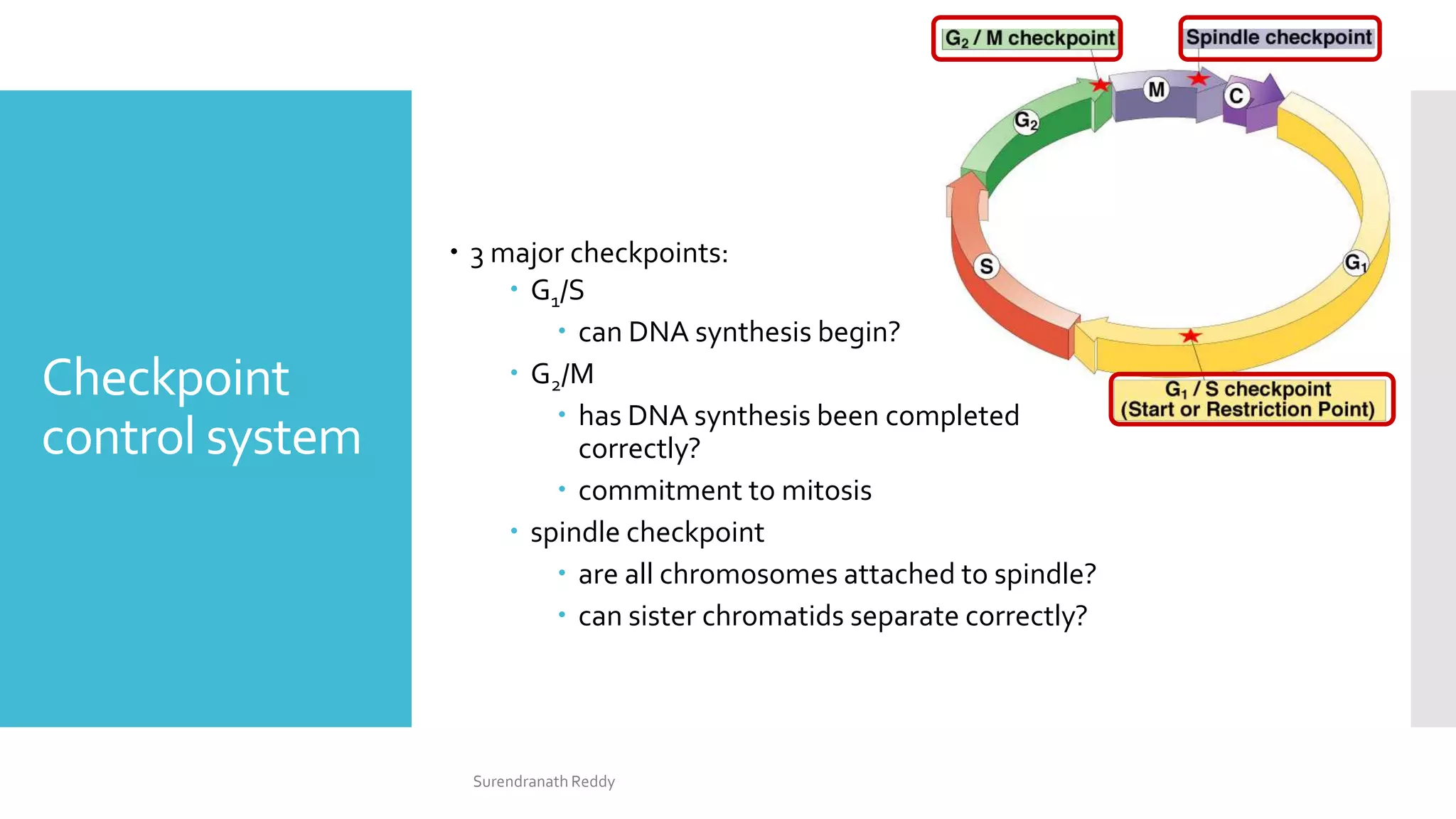
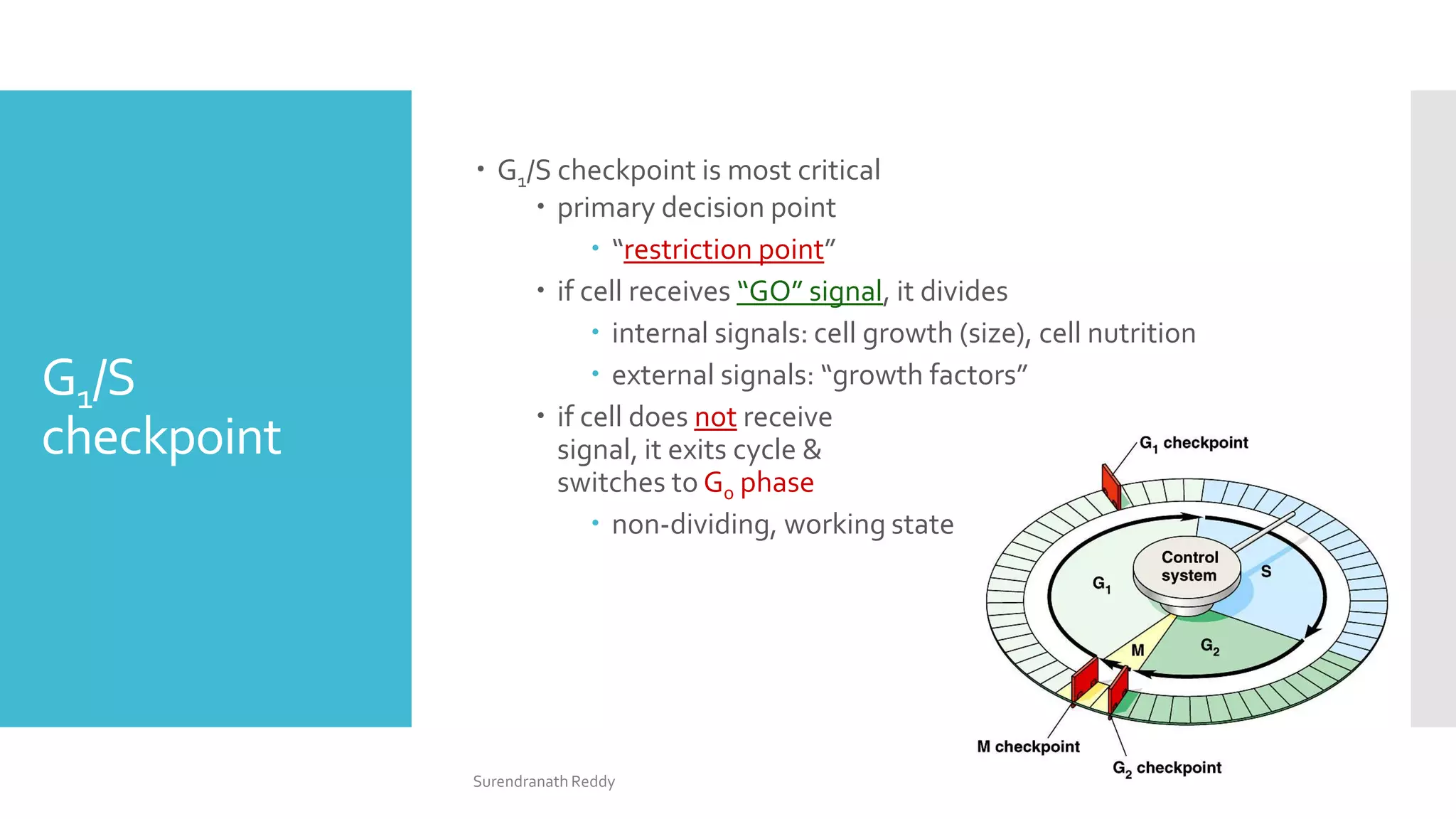
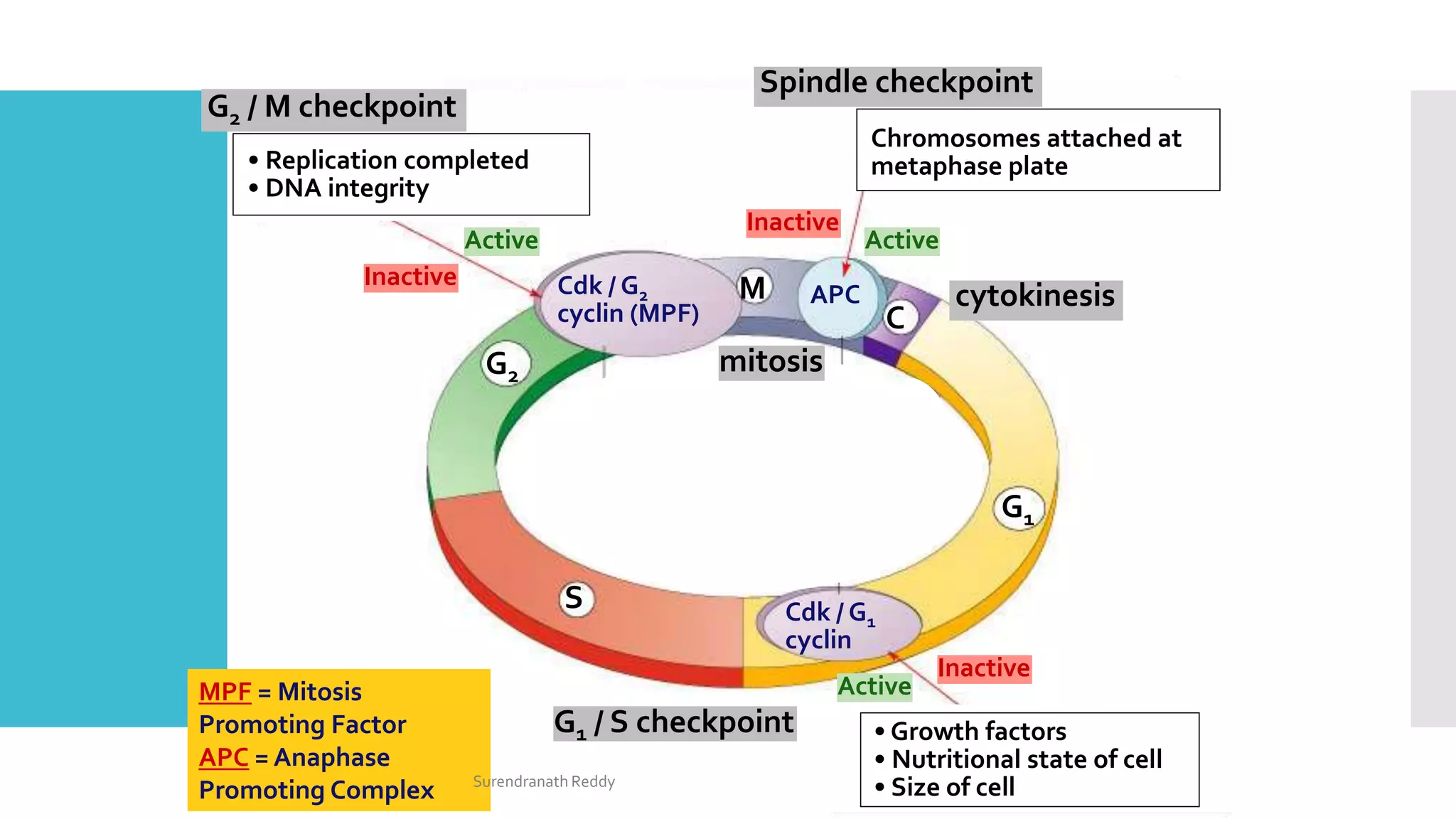
3) The cell cycle is regulated by checkpoints at the G1/S and G2/M transitions, which are controlled by complexes of cyclin-dependent kinases (CDKs) and cyclins. CDK-cyclin complexes phosphorylate proteins to drive the cell cycle forward past

















![CellCycle
Regulation
The cell cycle is regulated at the G1/S and G2/M boundaries
(checkpoints) by phosphorylation of complexes of a protein kinase
[cyclin-dependent kinase (Cdk) protein] and a cyclin (cytoplasmic
oscillator).
For example, the G2/M interface is regulated by M-Cdk complex
(formerly called Mitosis Promoting Factor, MPF), which is
responsible for the phosphorylation of spindle proteins, histones,
and lamins.
Phosphorylation of lamins results in their breakdown as well as the
dissolution of the nuclear envelope.There are different cyclins and
Cdks for each of the cell cycle checkpoints.
Overarching the Cdks are the Cdk inhibitors that form an
additional regulatory layer at each of the cell cycle checkpoints.
Study of the cell cycle is critical to an understanding of the
regulation of abnormal proliferation as occurs in cancer cells.
Surendranath Reddy](https://image.slidesharecdn.com/dnarepairmechanismscellcyclecheckpoints-211215100050/75/Dna-repair-mechanisms-amp-cell-cycle-checkpoints-25-2048.jpg)


