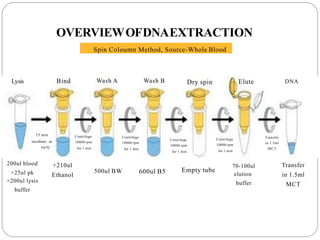
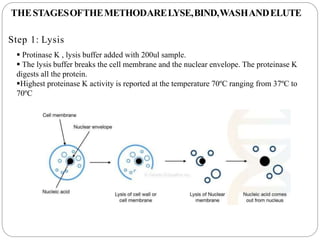
This document summarizes the spin column method for extracting DNA. It involves lysing cells to release DNA, binding the DNA to a silica membrane, washing to remove impurities, drying the membrane, and then eluting purified DNA. The key steps are lysing samples like blood to release DNA, binding DNA to a spin column, washing to remove proteins and other contaminants, drying the column, and then eluting purified DNA in buffer for storage or downstream applications like PCR or relationship testing.