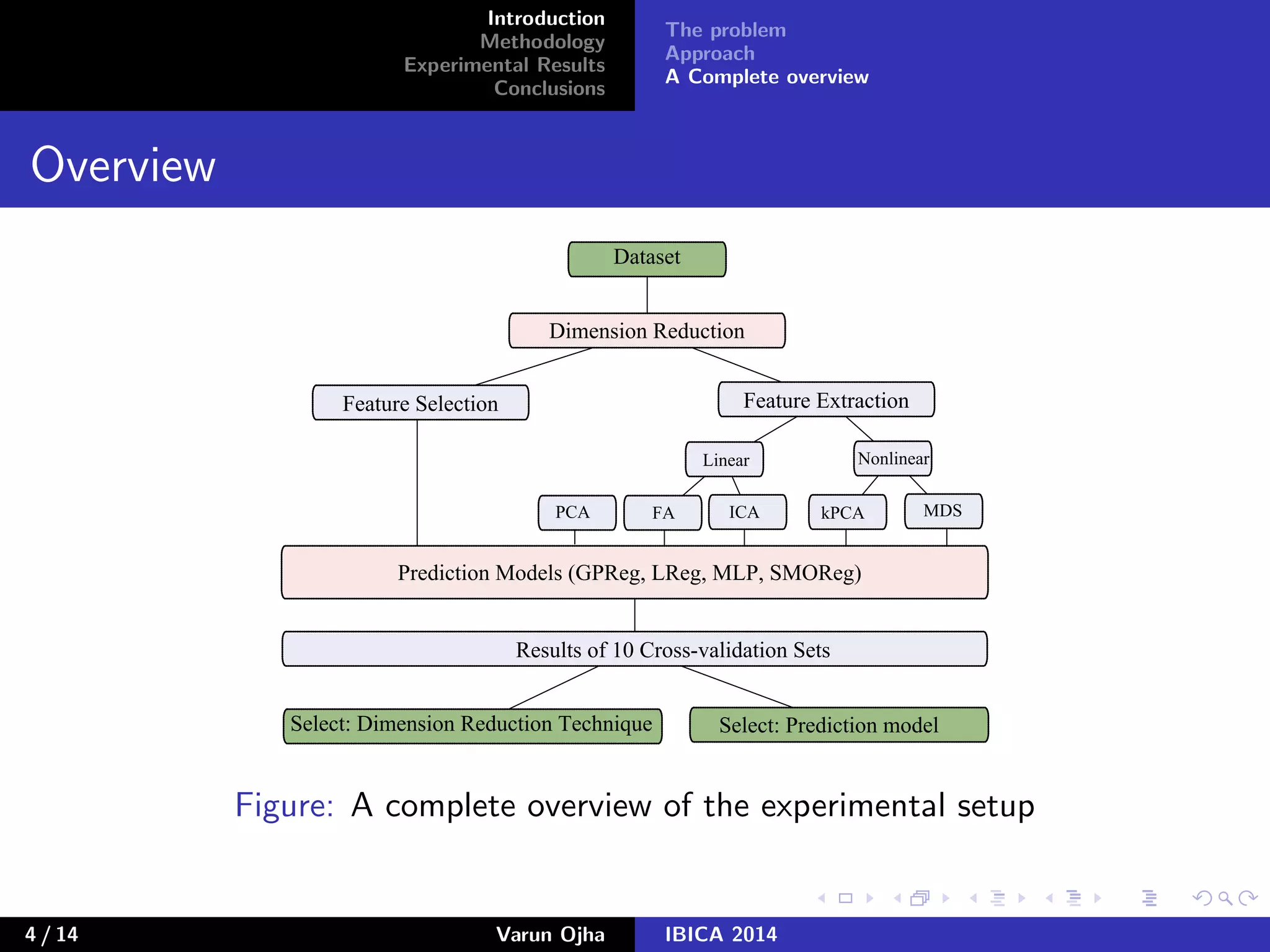
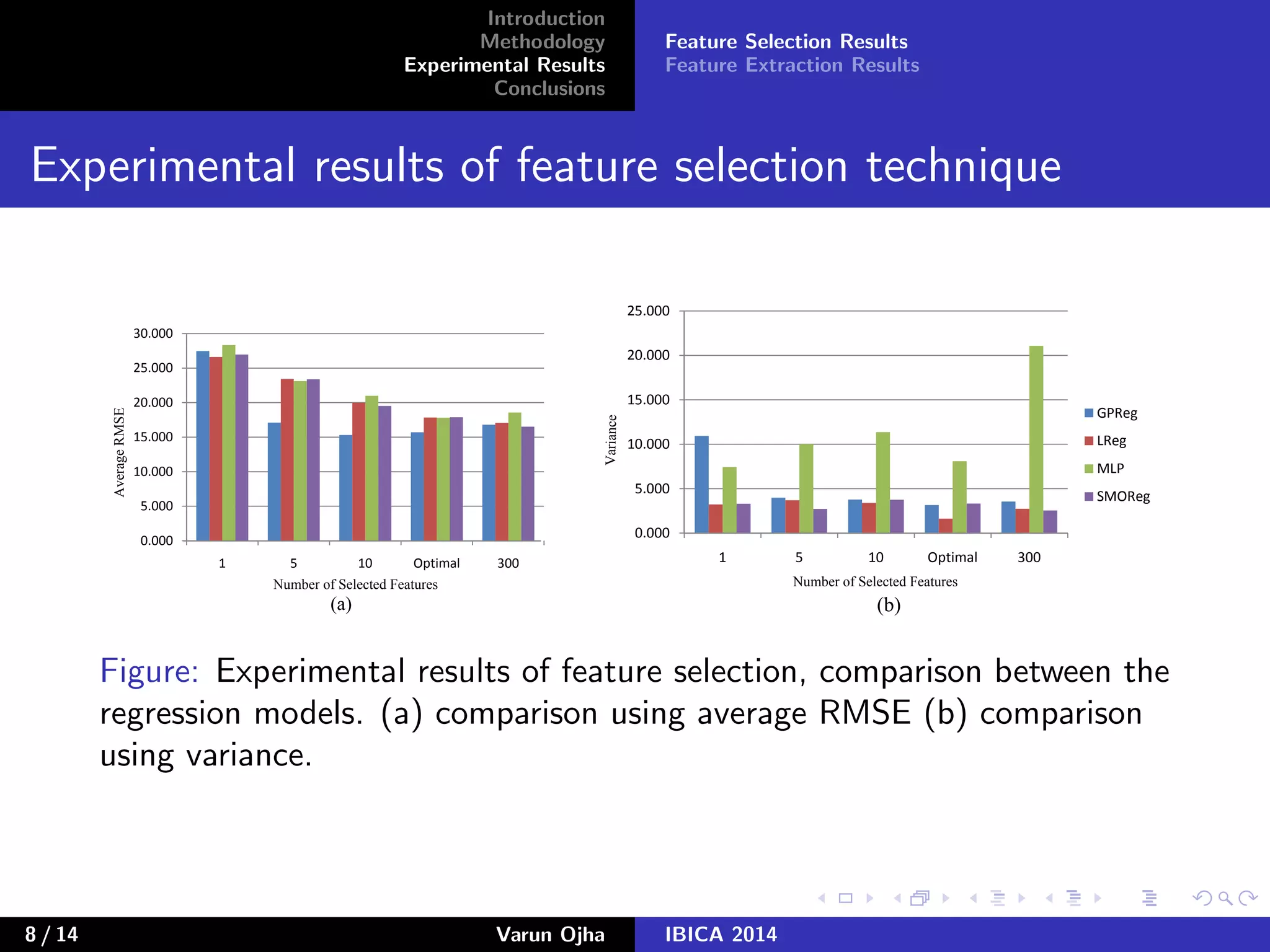
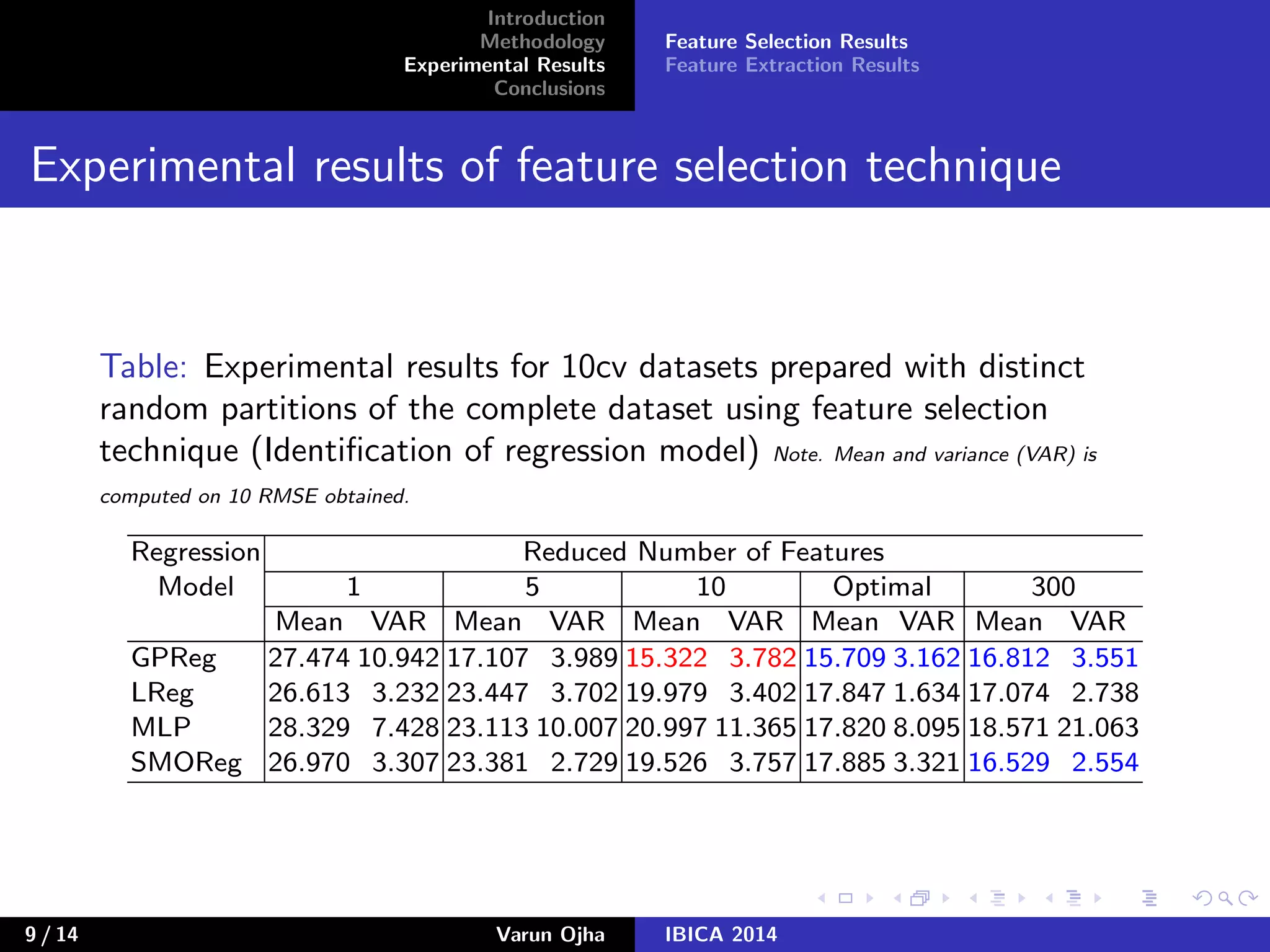
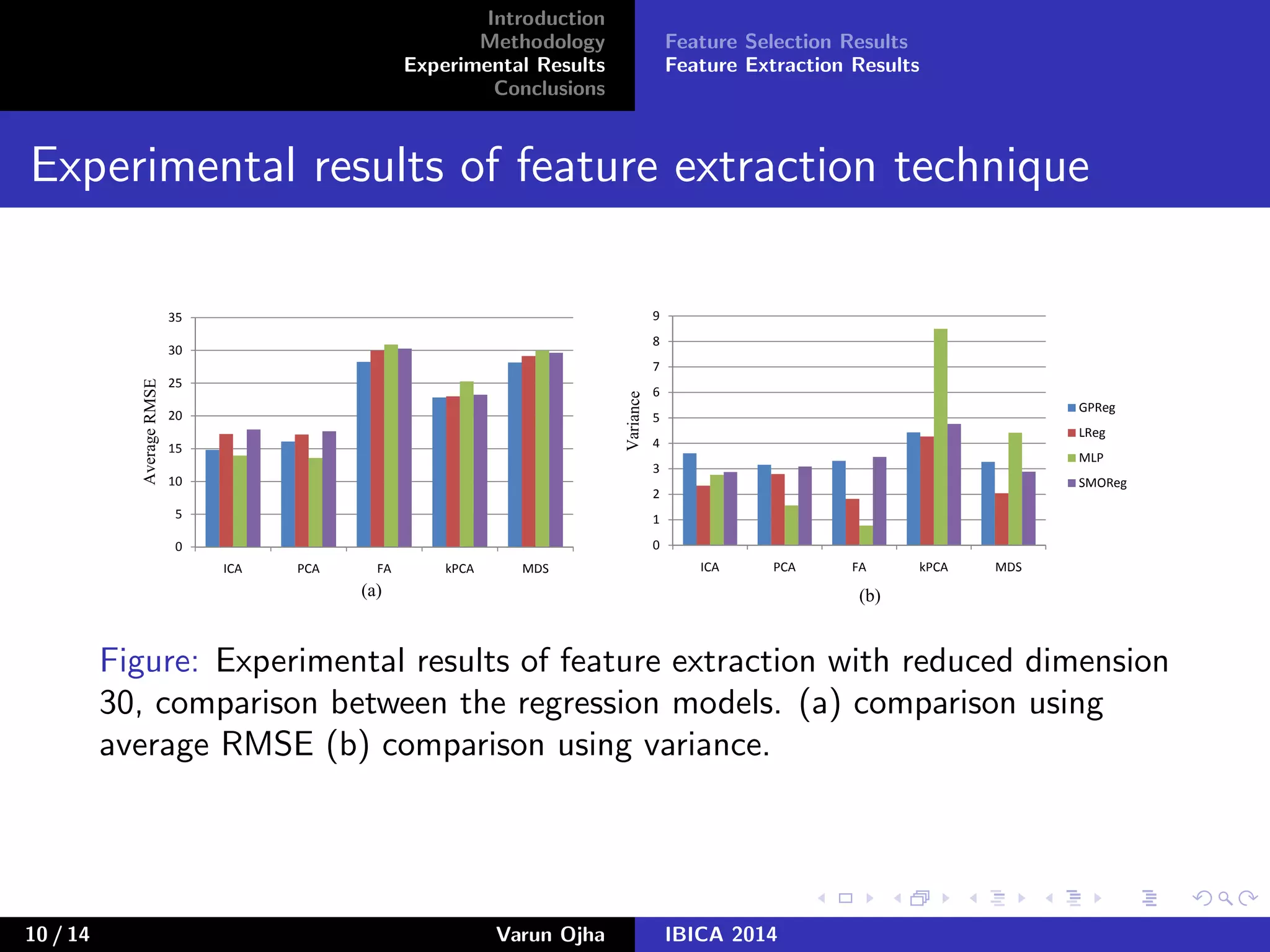
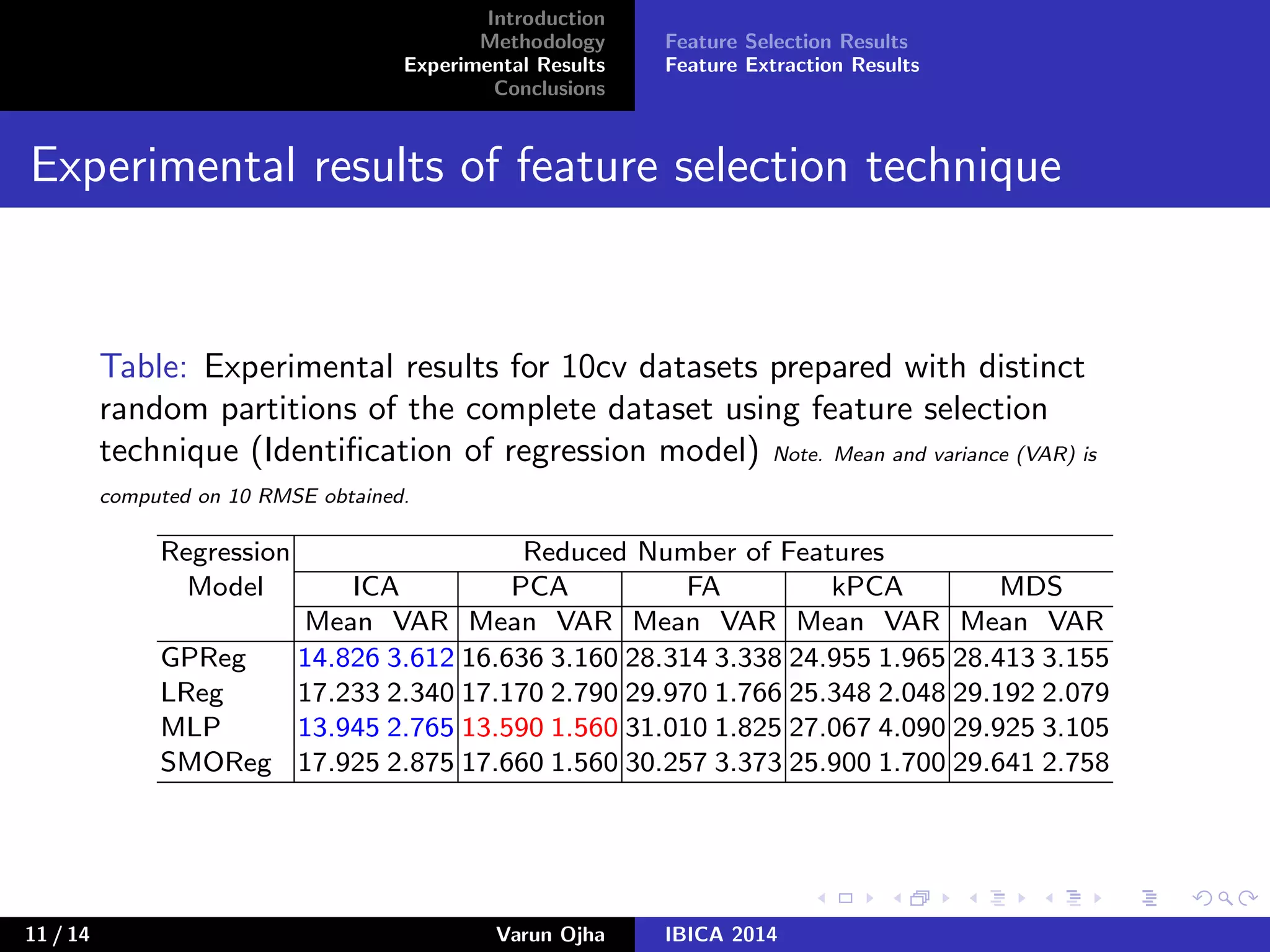
The document discusses dimensionality reduction and prediction of protein macromolecule dissolution profiles. It aims to reduce 300 input features to a smaller set using feature selection and extraction techniques. Experimental results show that Gaussian process regression with 10 selected features using backward feature elimination has the lowest error. For feature extraction, multilayer perceptron with dimensionality reduced to 30 using principal component analysis performed best. In conclusion, feature selection and extraction help improve prediction of dissolution profiles by reducing the number of input features.