Embed presentation
Download as PDF, PPTX





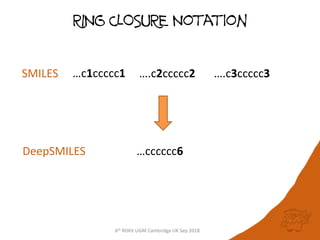
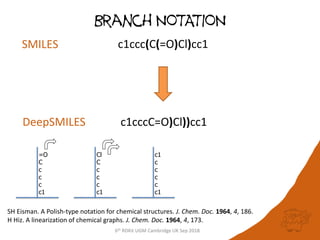


The document summarizes a presentation given at the 6th RDKit User Group Meeting in Cambridge, UK in September 2018. It discusses DeepSMILES, a modification of the SMILES notation for chemical structures that is designed for use in machine learning applications. DeepSMILES was presented by Noel O'Boyle and Andrew Dalke. The document provides examples of DeepSMILES notation for ring closures, branches, and installation instructions.







