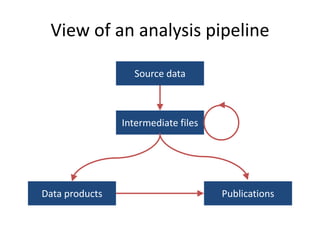
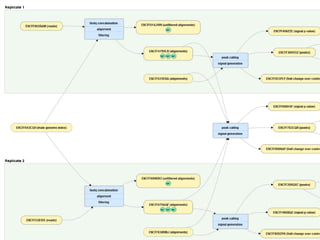
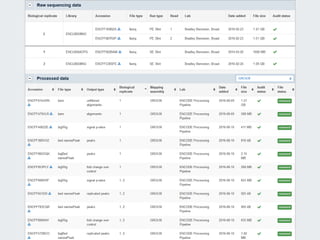
Michael M. Hoffman discusses the challenges researchers face with genomic data acquisition and distribution, including issues with data format, organization, and access. He proposes a solution where data must be archived and available at the time of publication, supported by a technical framework for data caching and economic considerations for central storage. The document highlights both practical and policy-related improvements needed in data management for effective research.