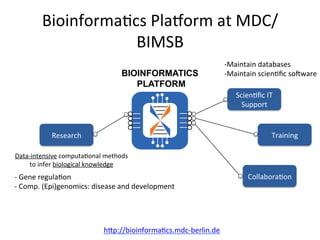
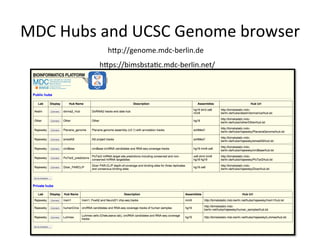
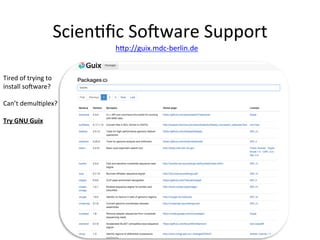
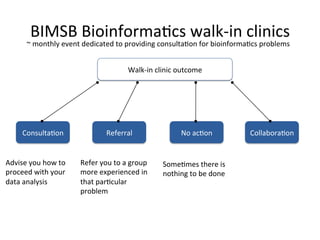
This document summarizes the Bioinformatics Platform at the Max Delbruck Center for Molecular Medicine in Berlin-Buch. The platform provides data analysis and computational resources to support genomic and epigenomic research. Services include maintaining databases and scientific software, providing scientific IT support, training courses, and collaborating with researchers on data analysis projects. Walk-in consultations are also offered monthly to help with bioinformatics problems.