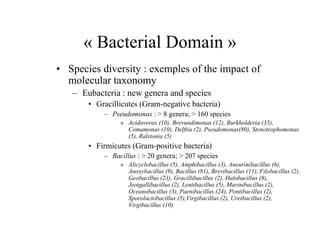
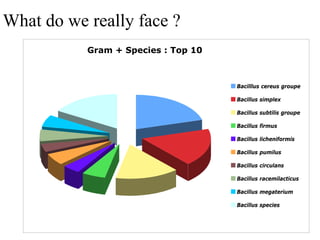
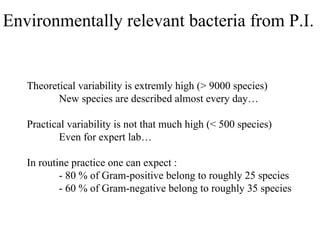
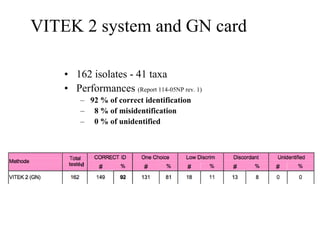
The document evaluates the performance of the new VITEK 2 system and identification cards for gram-positive (GP), gram-negative (GN) and Bacillus (BCL) bacteria commonly found in the environment of pharmaceutical industries. 404 bacterial isolates previously identified by 16S rRNA sequencing were tested. The GN card identified 92% correctly with 8% misidentified. The GP card identified 98.5% correctly with 1.5% misidentified. The BCL card identified 88.3% correctly, with 4.5% misidentified and 7.2% unidentified. The study demonstrates the VITEK 2 system can reliably identify the most common environmental bacteria isolated from pharmaceutical industries.