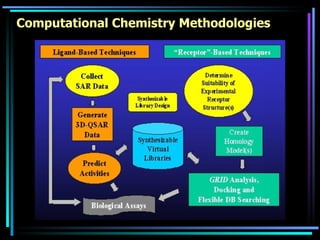
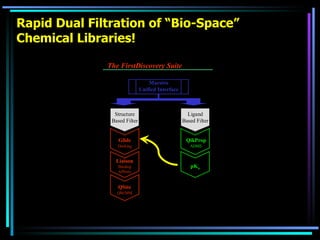
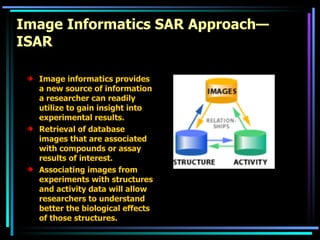
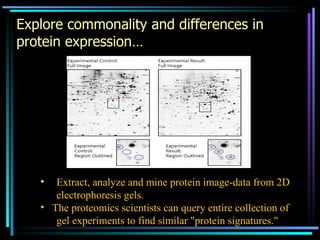
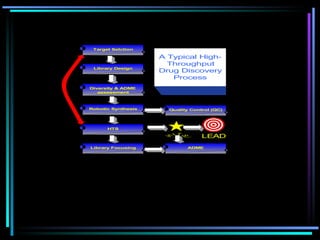
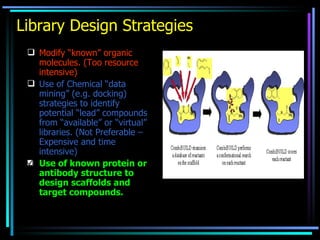
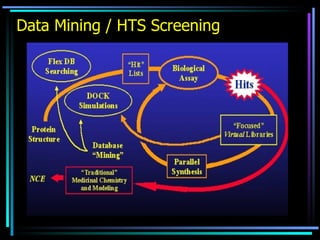
The document discusses strategies for efficiently designing "bio space" chemical libraries to identify drug molecules. It proposes using computational chemistry to integrate data from various R&D departments and design libraries with drug-like properties. Building blocks and scaffolds will be selected from existing drugs based on properties like absorption, distribution, metabolism, and excretion. This approach aims to design focused libraries more likely to yield meaningful hits in high-throughput screening that are inherently more drug-like.





![High Valued New Chemical Entities Approx. 40% drugs in clinical trials are discarded because they do not show the correct adsorption, distribution, metabolism, and excretion properties (ADME) [Drug Discovery Today 1997, 2, 436]. Results? Loss of hundreds of Millions of dollars.](https://image.slidesharecdn.com/biospacelibraries-12736227227264-phpapp02/85/Biospace-Libraries-6-320.jpg)