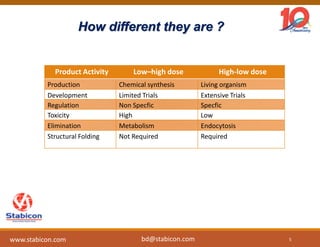
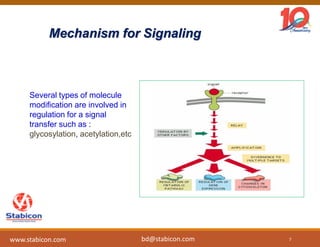
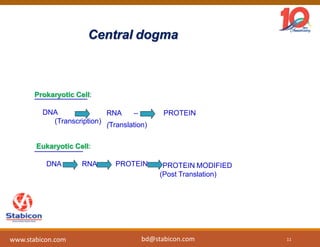
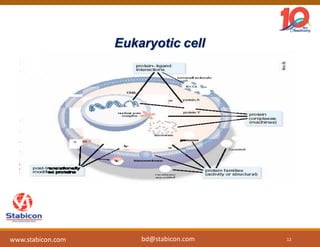
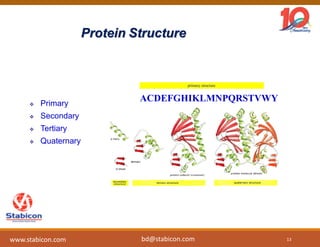
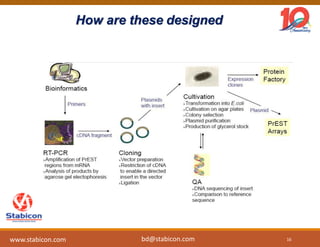
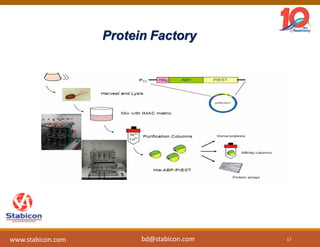
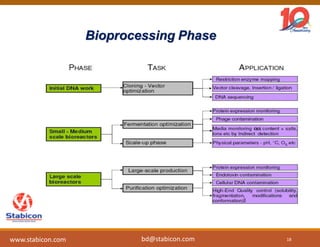
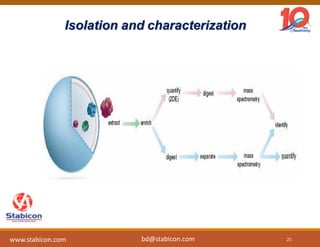
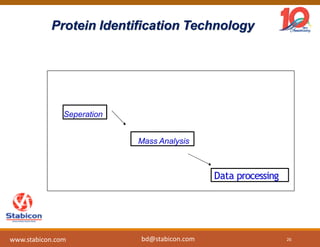
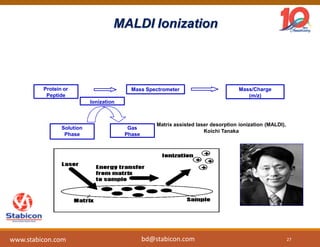
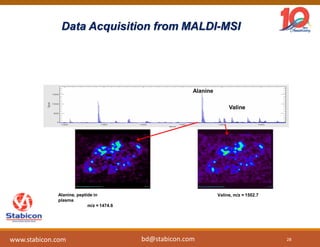
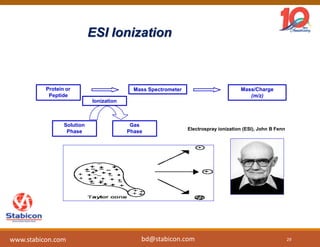
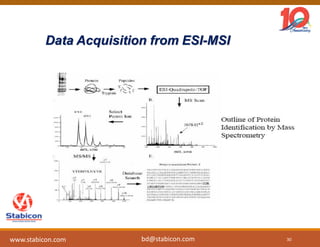
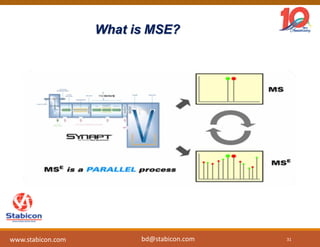
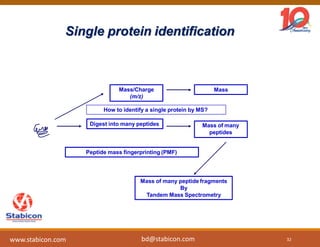
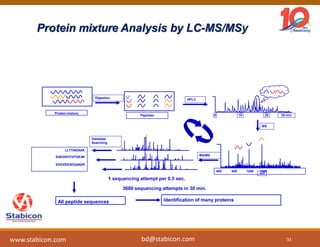
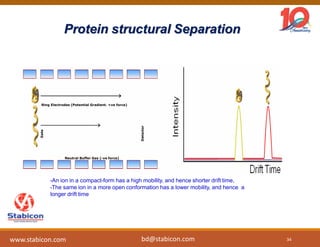
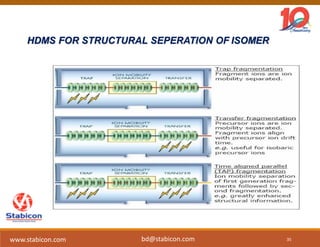
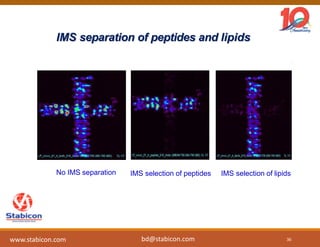
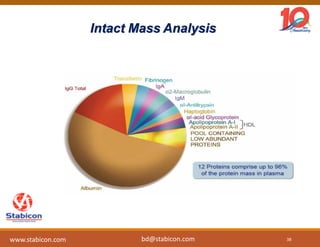
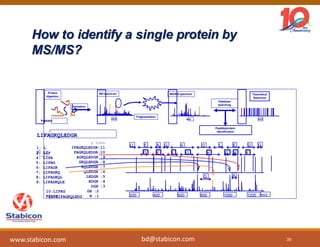
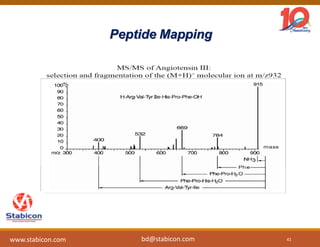
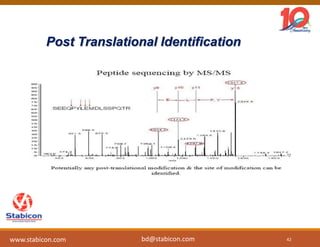
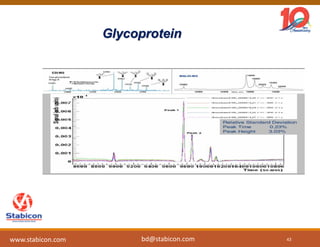
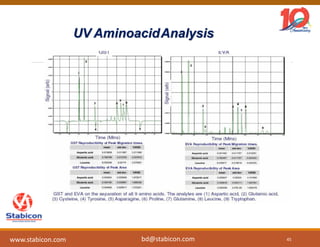
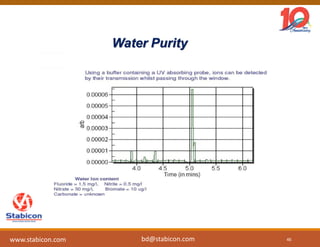
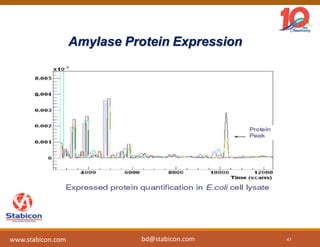
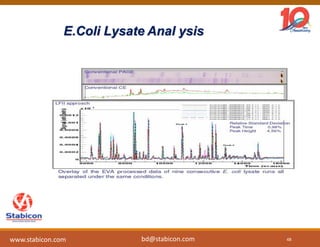
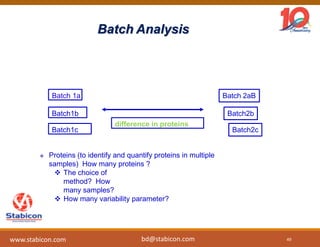
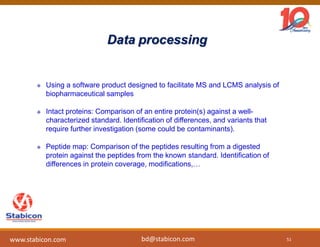
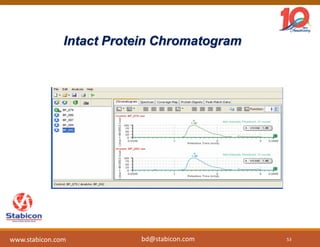
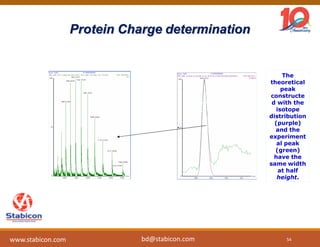
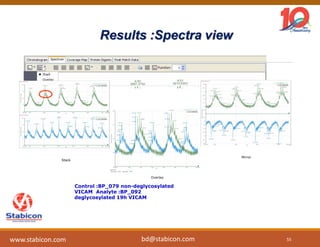
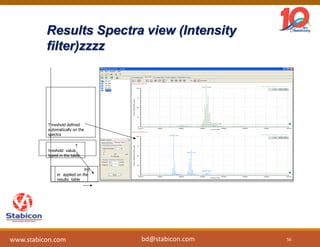
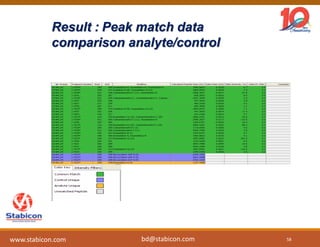
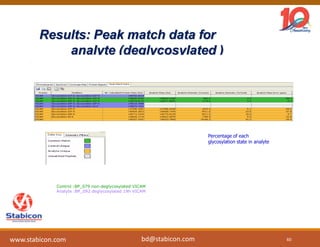
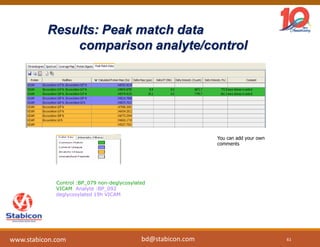
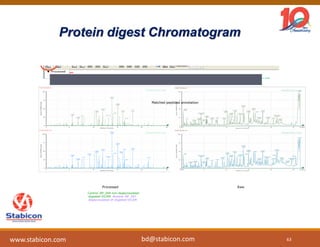
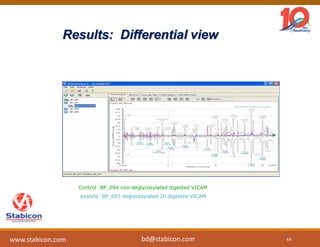
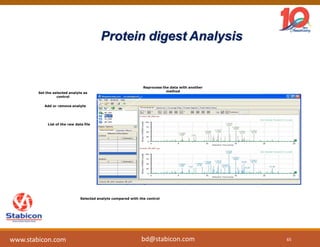
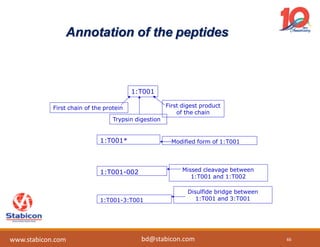
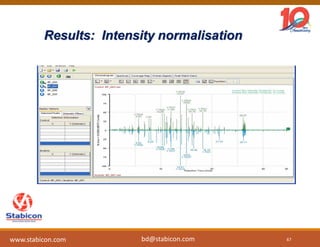
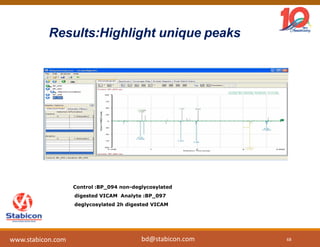
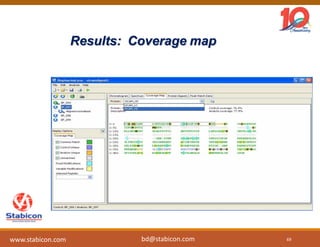
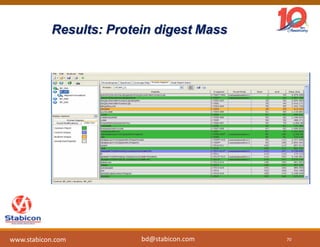
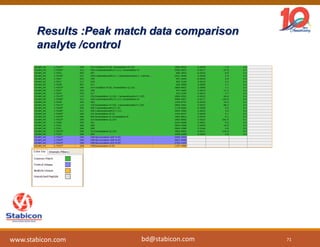
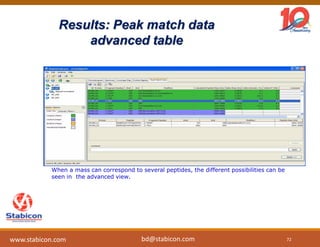
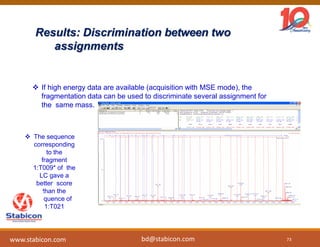
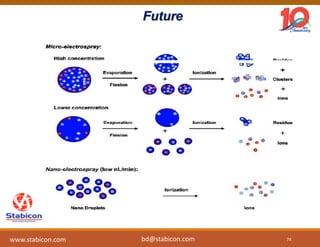
The document discusses the production, characterization, and monitoring of biologics, which are large therapeutic proteins significantly larger than traditional small molecules. It outlines methods used for their identification and characterization, including mass spectrometry and data processing techniques. Additionally, it highlights the unique properties of biologics, such as their signaling mechanisms and regulatory requirements, as well as examples of marketed biologics.