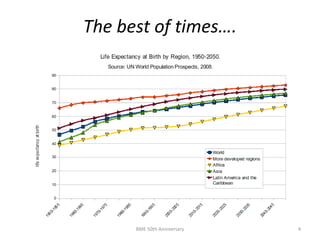
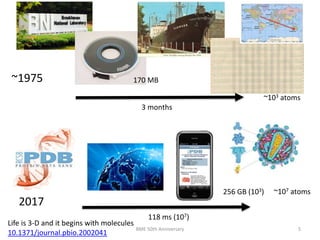
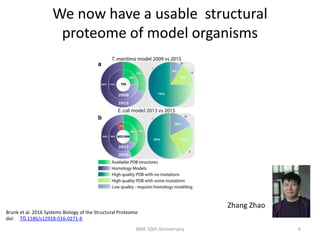
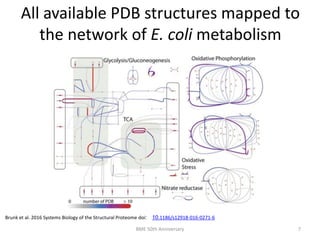
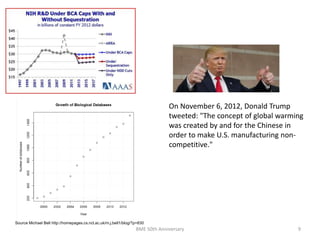
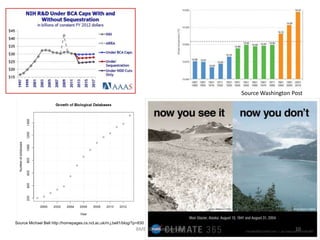
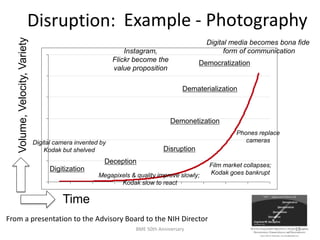
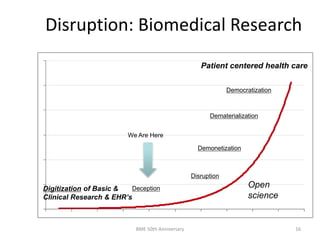
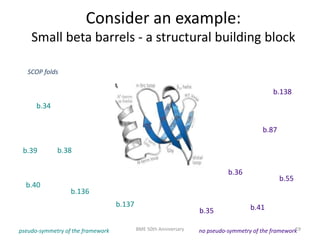
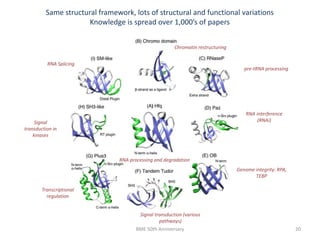
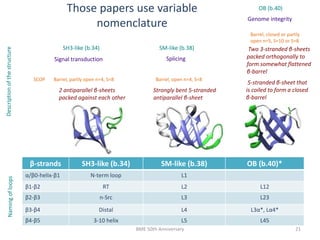
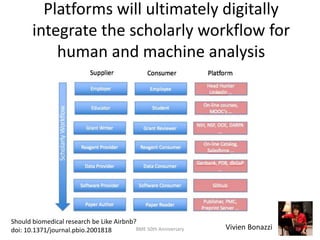
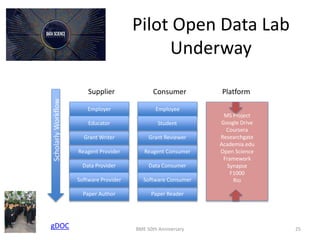
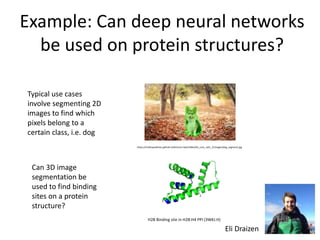
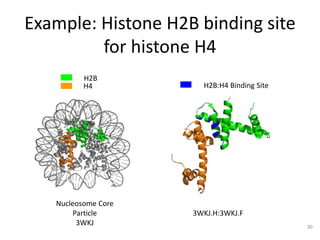
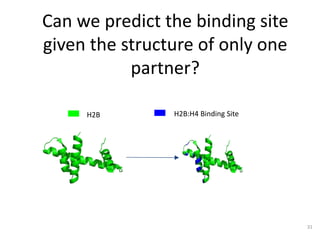
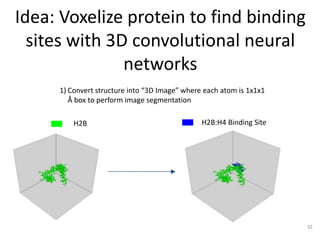
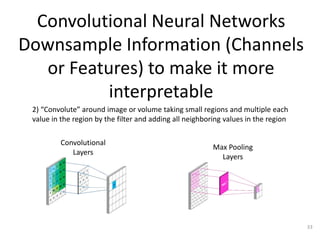
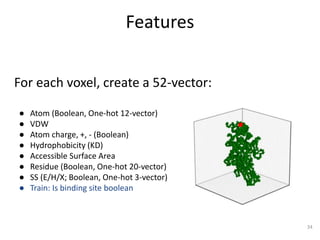
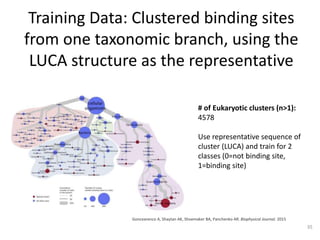
The document discusses the evolution and future prospects of biomedical engineering, reflecting on 50 years of progress and emphasizing the need for good scientific practices. It highlights the potential of digital platforms and AI in transforming research methodologies and data management in the field. The author calls for collaboration and innovation to drive meaningful changes in biomedical research moving forward.