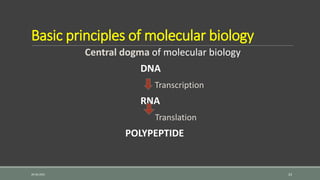
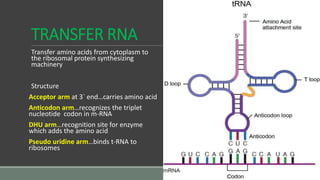
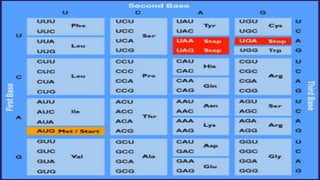
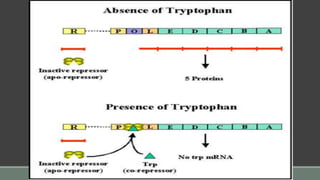
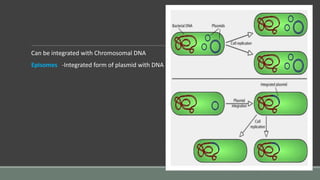
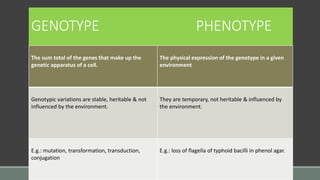
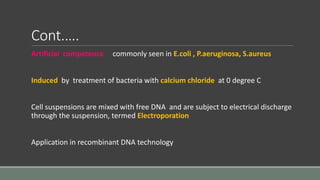
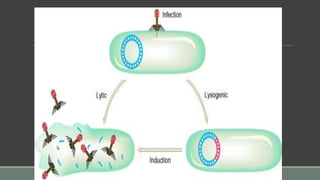
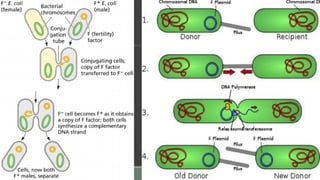
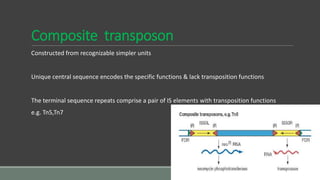
The document provides an overview of bacterial genetics, including foundational concepts like the central dogma (DNA to RNA to protein), mechanisms of gene transfer (transformation, transduction, conjugation), and the structure and function of various genetic elements such as plasmids and transposons. It discusses key historical figures in genetics and critical processes like DNA replication, transcription, and translation, highlighting the importance of mutations and gene regulation in bacterial adaptability. Additionally, it covers the implications of mobile genetic elements and their role in antibiotic resistance.