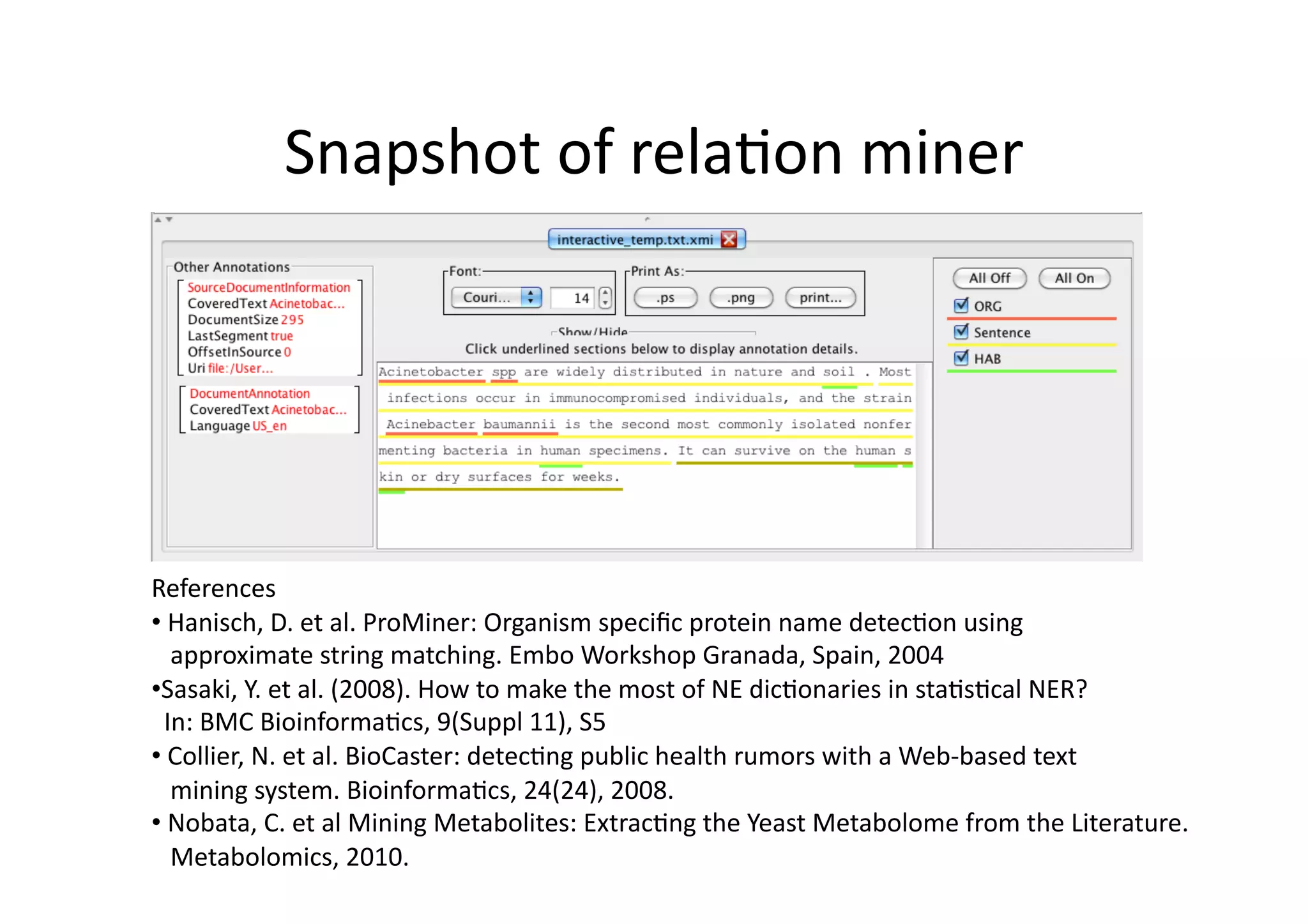
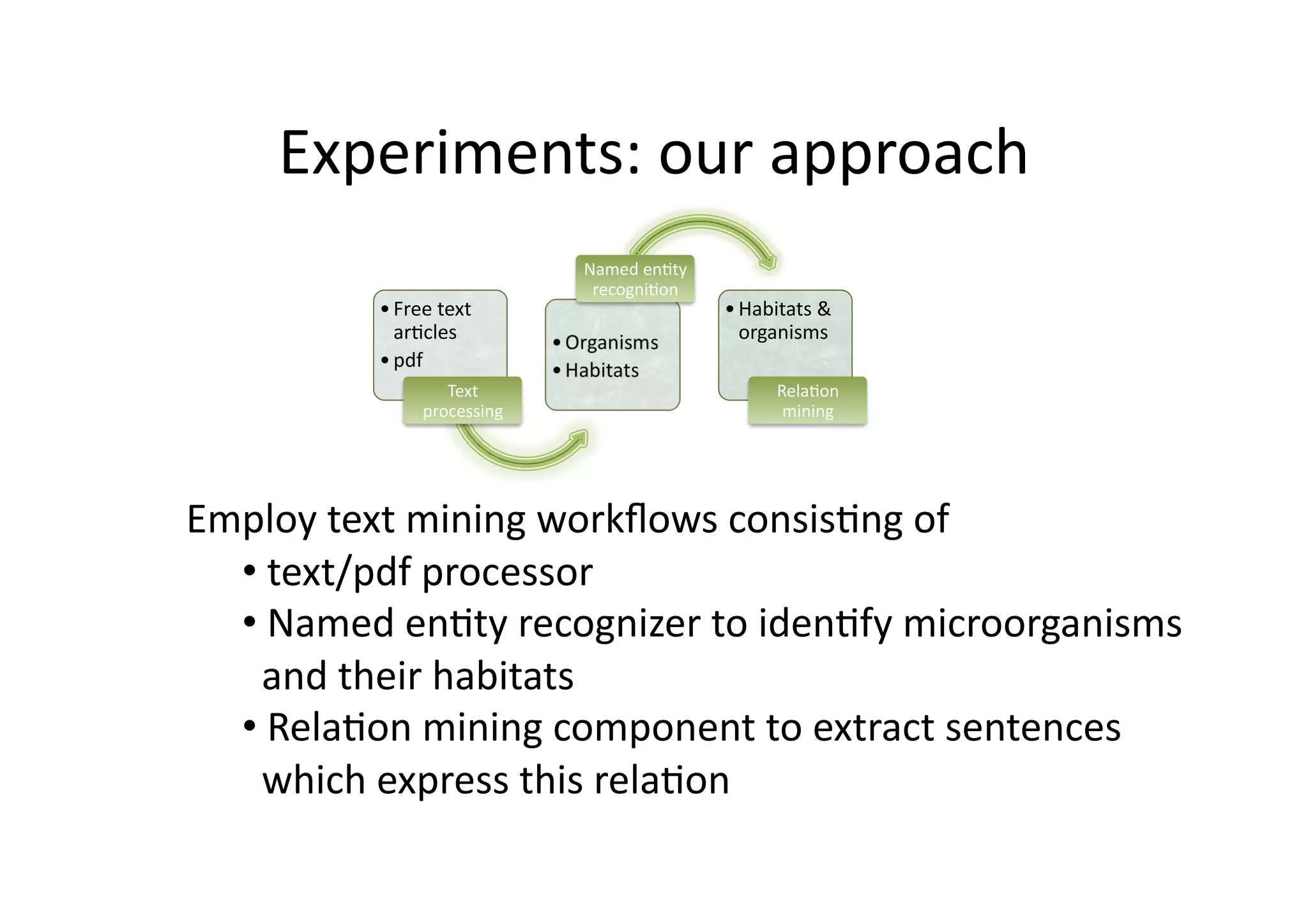
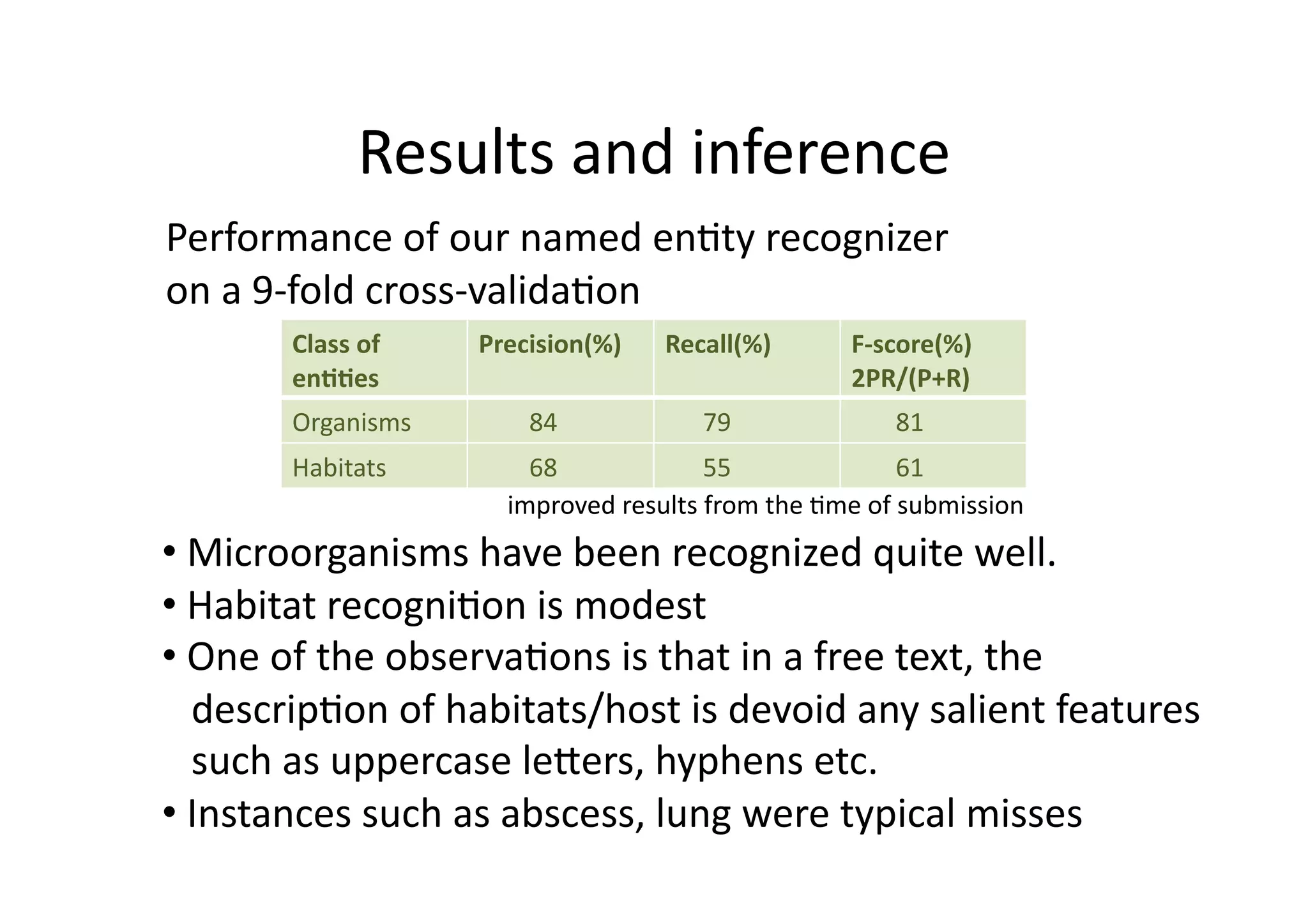
This document summarizes an approach to automatically extract microorganisms and their habitats from free text using text mining workflows. The approach uses a named entity recognizer combining dictionaries and machine learning to identify organisms and habitats. It then employs relation mining to extract sentences expressing relationships between organisms and habitats. Evaluation shows the organism recognition achieves 84% precision but habitat recognition is lower at 68% precision, limiting the relation mining performance. Ongoing work aims to improve the habitat model and make the overall approach more robust to noise from PDF to text conversion.



![Experiments:
relevant
work
• Iden'fica'on
of
named
en''es
such
as
microorganisms,
diseases,
genes
etc.,
has
received
sufficient
importance
from
the
scien'fic
community
at
large
[Sasaki,
Hanisch,
Chikashi]
• Researchers
have
also
used
ontology
based
approaches
to
iden'fy
concepts
such
as
public
health
rumors
etc
[Biocaster]](https://image.slidesharecdn.com/nactemib2011-110527050030-phpapp02/75/Automatic-extraction-of-microorganisms-and-their-habitats-from-free-text-using-text-mining-workflows-4-2048.jpg)
![Experiments:
our
approach
• The
named
en'ty
recognizer
used
a
hybrid
dic'onary-‐machine
learning
based
approach
– It
combined
the
informa'on
dic'onaries
with
a
feature
set
for
a
condi'onal
random
field
(CRF)
based
classifier
[Mallet]
– The
CRFs
used
a
linear
chain
model
and
were
trained
on
a
corpus
consis'ng
of
32
full
papers](https://image.slidesharecdn.com/nactemib2011-110527050030-phpapp02/75/Automatic-extraction-of-microorganisms-and-their-habitats-from-free-text-using-text-mining-workflows-6-2048.jpg)



![Discussion
• Pdf-‐to-‐text
sentence
examples
These
mechanisms
may
have
evolved
in
bacterial
pathogens
to
increase
the
frequency
of
phenotypic
varia'on
in
genes
involved
in
1
100,000
200,000
300,000
1,600,00
Figure
2
Circular
representa'on
of
the
H.
pylori
26695
chromosome.
[Clearly,
data
from
a
table
and
figure
corrupted
the
sentence]
airborne
pigs
[noisy
conversion
of
table
discussing
airborne
diseases
in
pigs
]](https://image.slidesharecdn.com/nactemib2011-110527050030-phpapp02/75/Automatic-extraction-of-microorganisms-and-their-habitats-from-free-text-using-text-mining-workflows-11-2048.jpg)