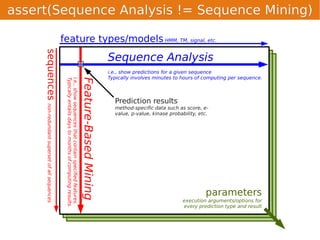
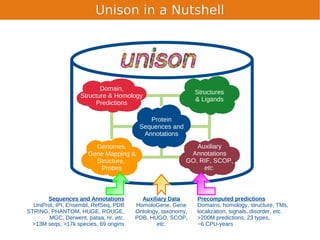
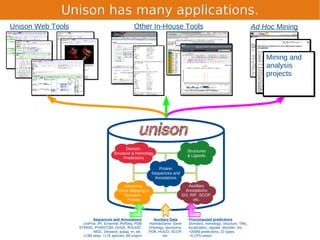
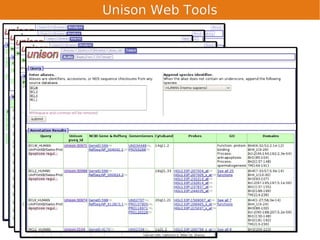
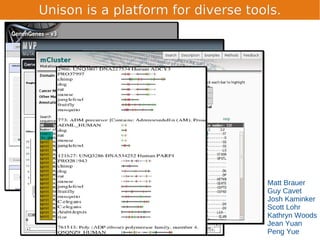
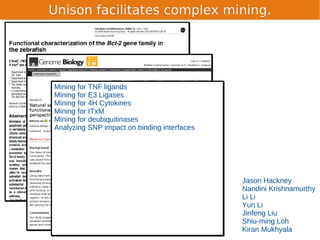
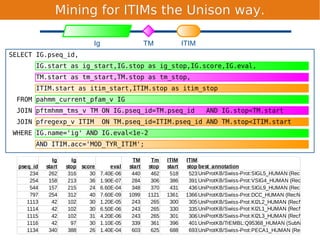
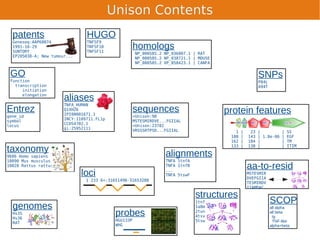
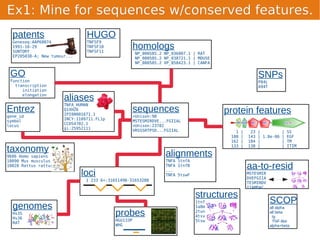
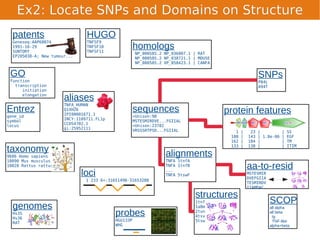
Unison is an integrated platform for computational biology discovery freely available at http://unison-db.org/. It contains over 13 million protein sequences, precomputed predictions for over 200 million sequences across 23 feature types, auxiliary data like GO terms and structures, and tools for complex queries and mining of sequences with specified features. Some examples of mining include finding sequences with immunoglobulin domains, transmembrane domains, and ITIM domains, or locating SNPs and domains on protein structures. Unison facilitates diverse computational biology tasks through its comprehensive integrated data and ability to easily perform complex queries and analysis.