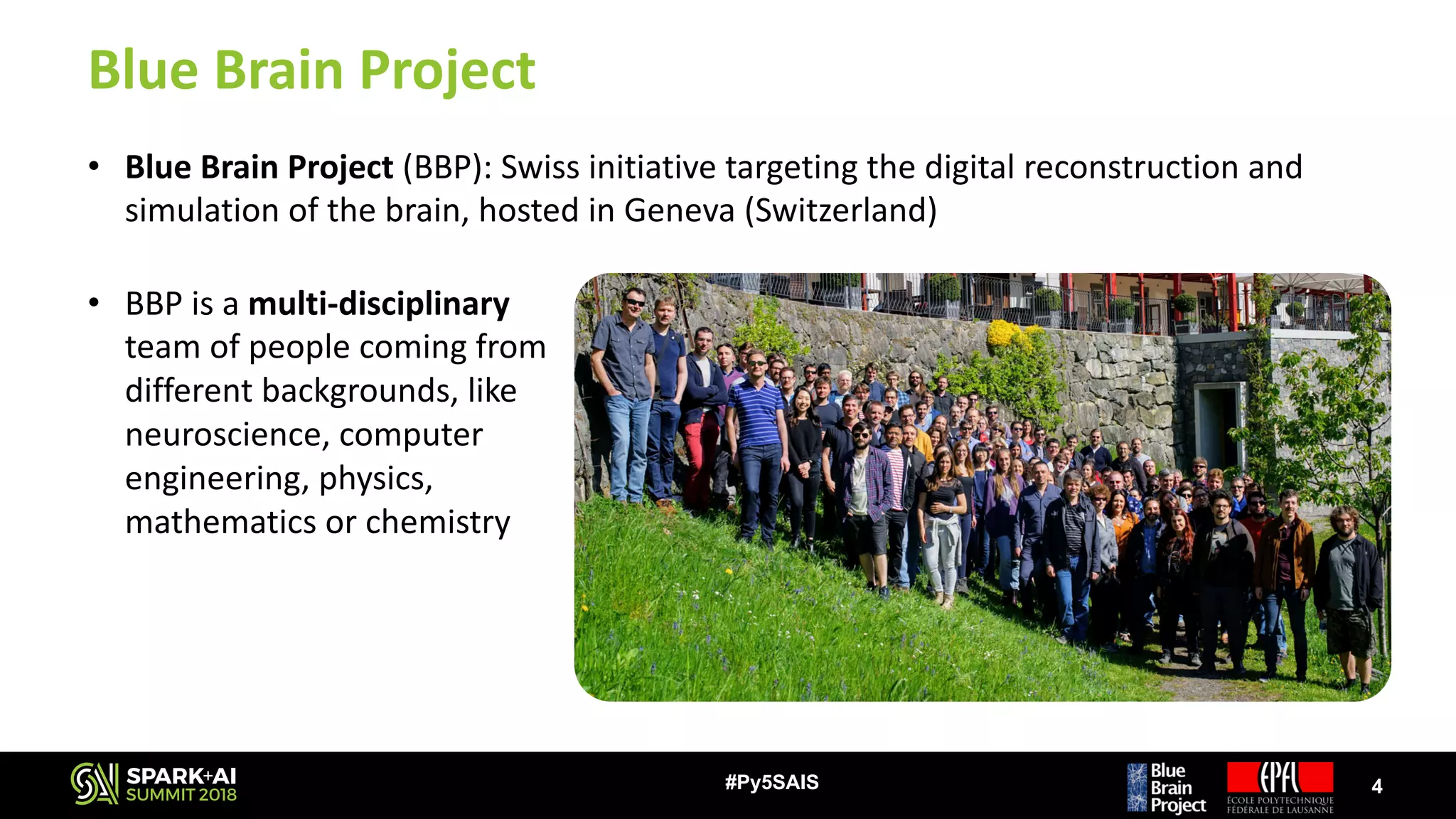
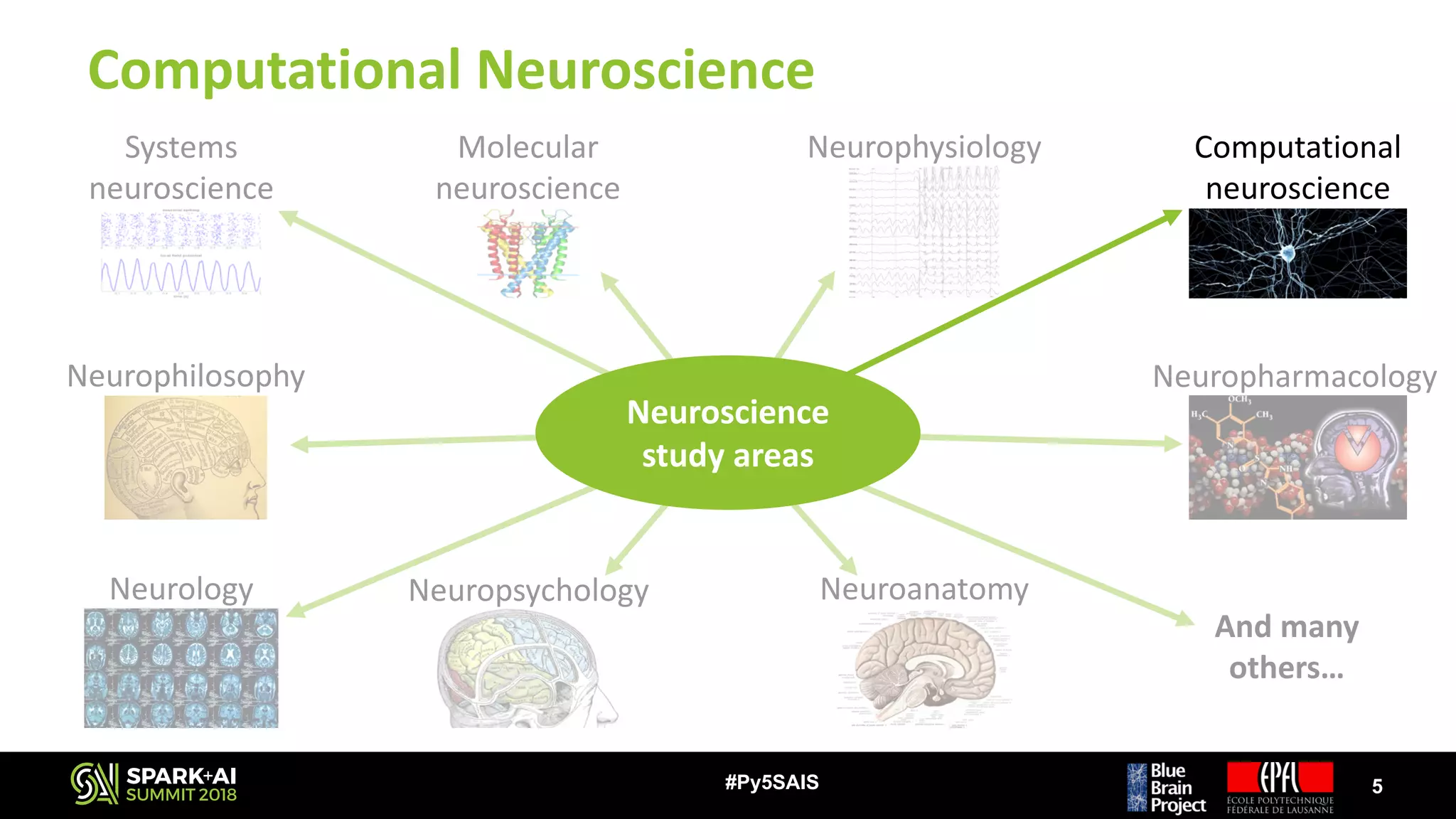
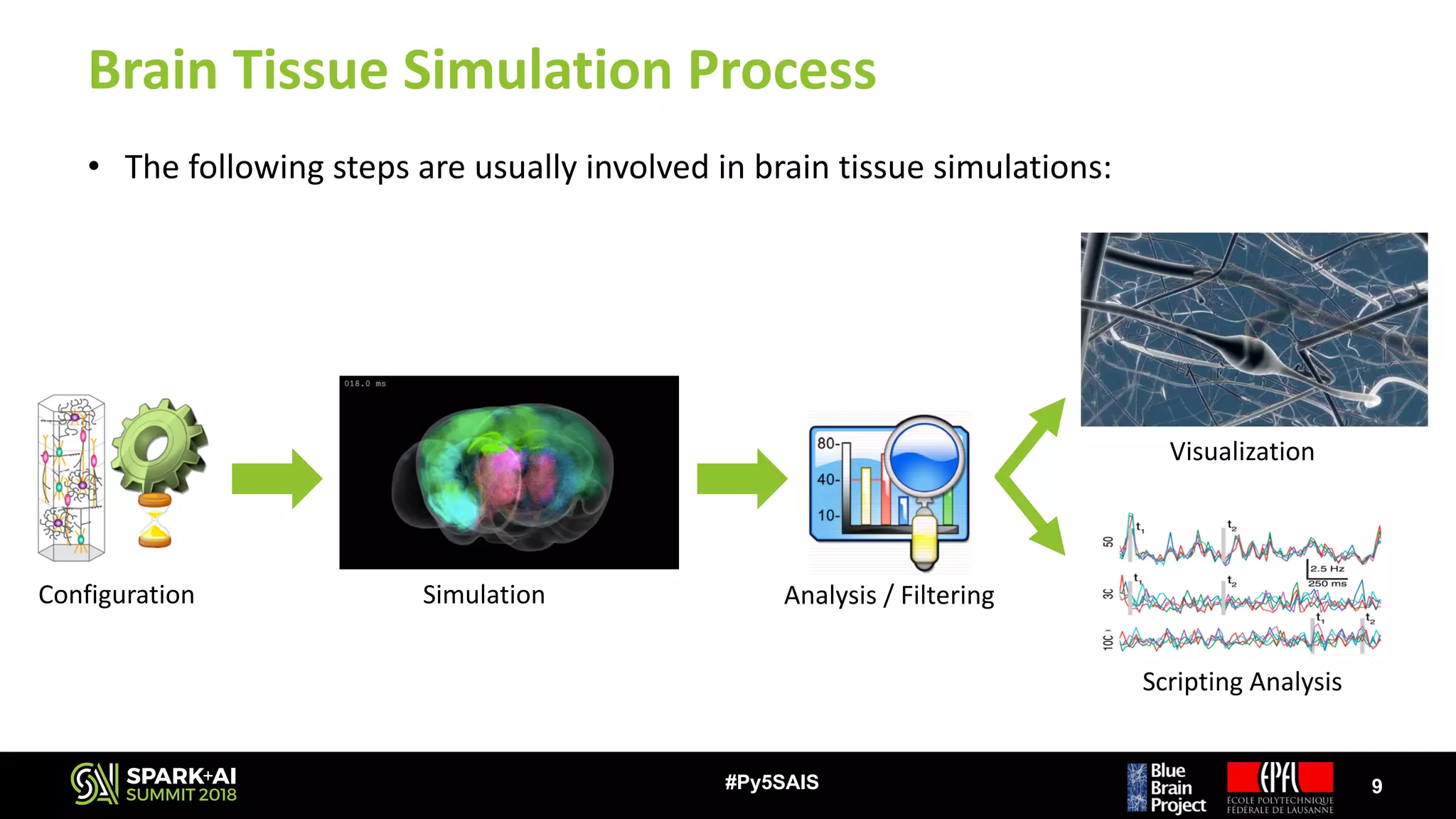
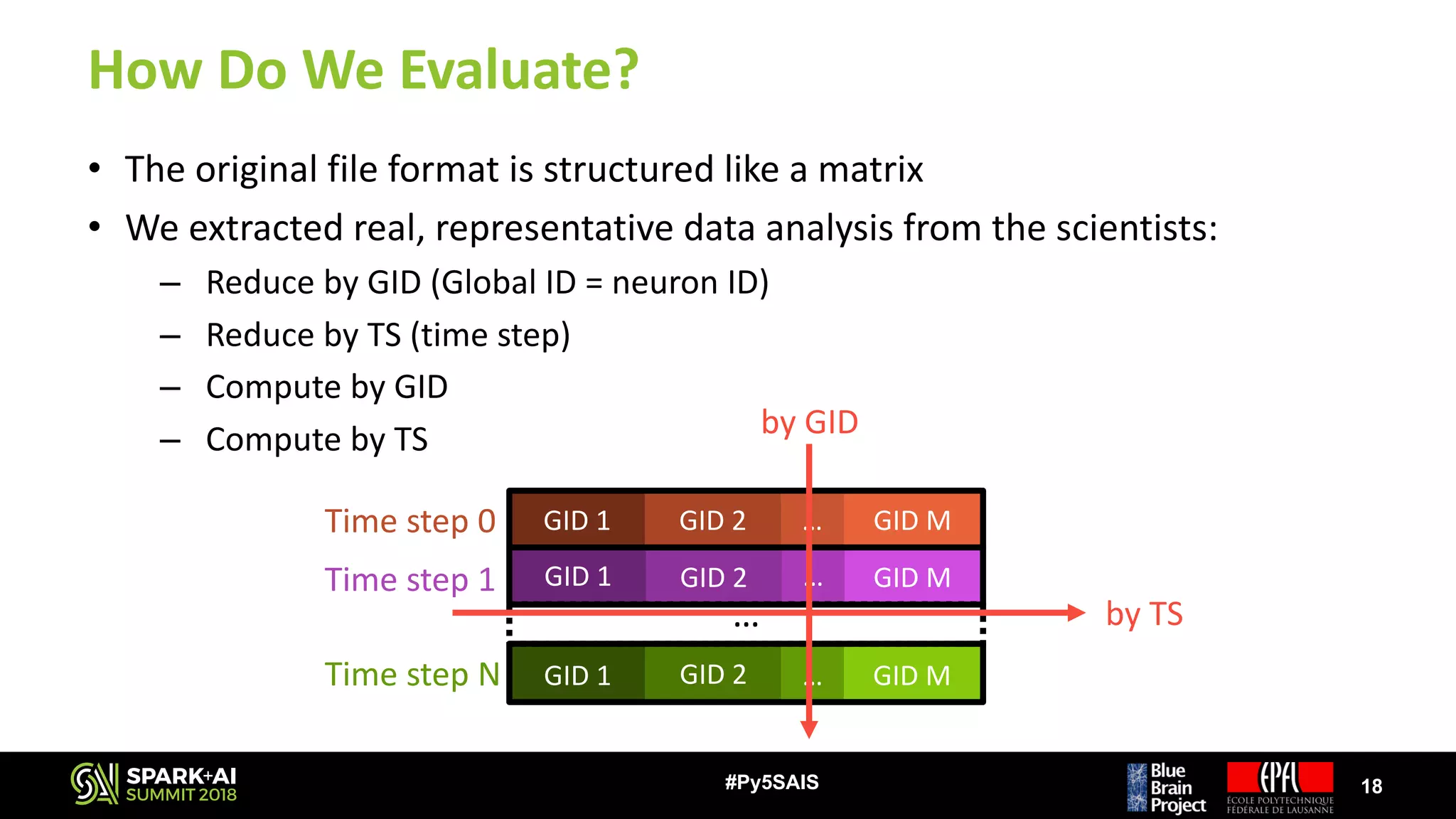
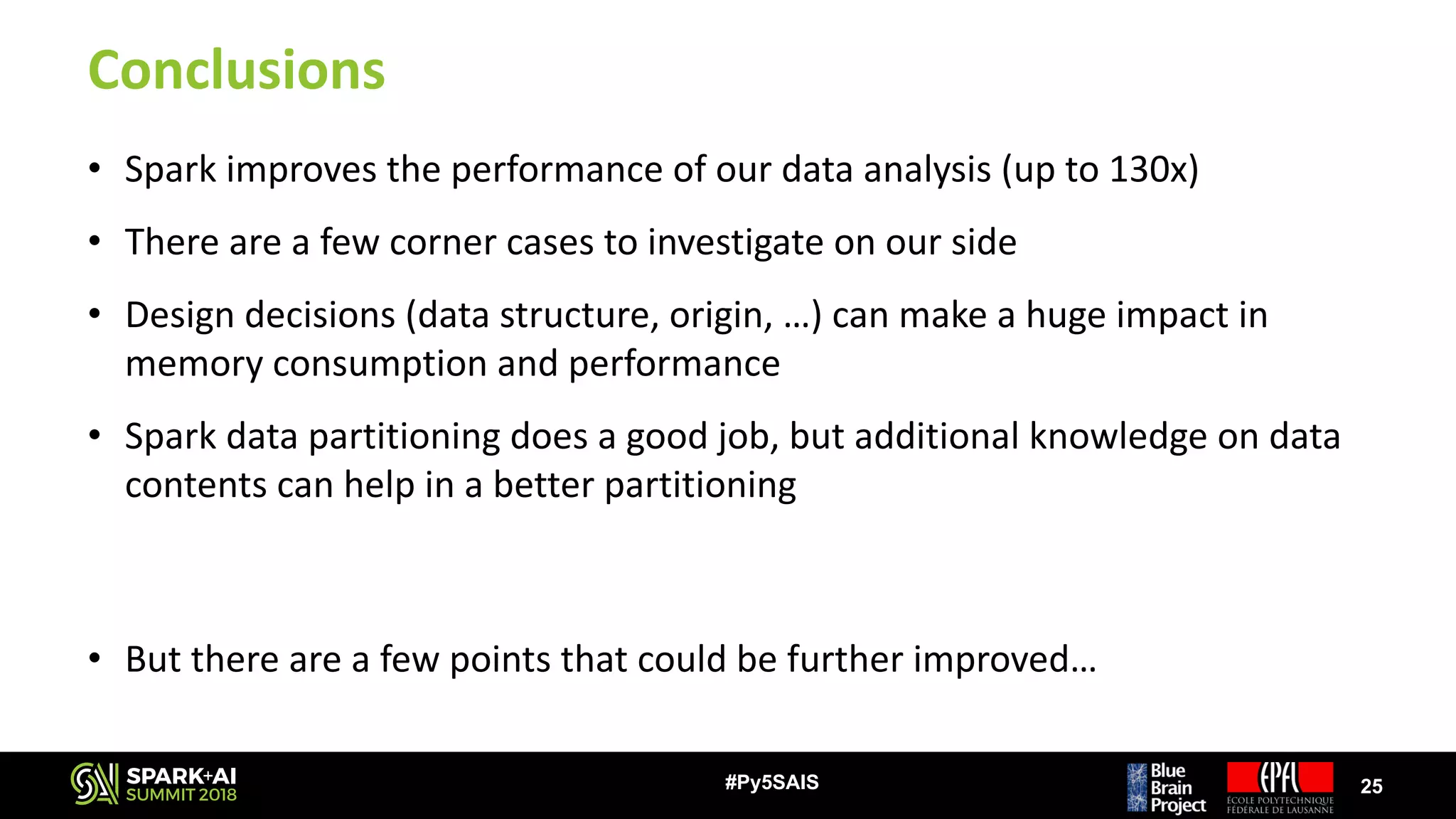
The document discusses the Blue Brain Project (BBP), a Swiss initiative aimed at digitally reconstructing and simulating the brain to aid in neuroscience research. It highlights the challenges of analyzing large simulation datasets and proposes using Apache Spark to improve data analysis performance, achieving speed-ups of up to 130 times. The analysis explores various data structures and their impact on memory consumption and performance, concluding that Spark can significantly enhance efficiency in processing brain simulation data.
![BBP’s Target Contributions to Neuroscience
• Help scientists understand how the brain works internally
• Recently, BBP has been able to reproduce the electrical behavior
of a neocortex fragment by means of a computer reconstruction[1]
– Brain volume: 1/3 mm3
– 30’000 neurons
– 40 million synapses (connections between neurons)
– This model has revealed novel insights into the functioning of the neocortex
• Supercomputer-based simulation of the brain provides a new tool to study the
interaction within the different brain regions
• Understanding the brain not only will help the diagnosis and treatment of brain
diseases, but also will contribute to the development of neurorobotics,
neuromorphic computing and AI
6#Py5SAIS](https://image.slidesharecdn.com/5juditplanas-180618234325/75/Accelerating-Data-Analysis-of-Brain-Tissue-Simulations-with-Apache-Spark-with-Judit-Planas-6-2048.jpg)
![Simulation Neuroscience at Different Levels
7#Py5SAIS
Model Point neurons
Morphologicallydetailed
neurons
Subcellular (molecular)
Simulator
Example
NEST
Spikingneural
networks,focused
ondynamics,size
andstructure
NEURON
Supports cells with
complexanatomical
andbiophysical
properties
STEPS
Detailedmodels of
neuronalsignaling
pathways at
molecularlevel
HW Platform Full scale Ksupercomp. 20 racks BG/Q Full scale on Sango
HumanBrain% 2.3 % 0.1 % 1.2 x 10-9 %
# Neurons 1.86 x 109 82 x 106 1
BiologicalTime 1 s 10 ms 30 ms
SimulationTime 3275 s 220 s 26.8 s
Sim TimeNorm.:
1 neuron, 1 thread,
1 s biotime
0.003 ms 0.28 ms 1.3 x 106 s
All these values are approximate and should NOT be used for technical purposes [2, 3, 4]](https://image.slidesharecdn.com/5juditplanas-180618234325/75/Accelerating-Data-Analysis-of-Brain-Tissue-Simulations-with-Apache-Spark-with-Judit-Planas-7-2048.jpg)

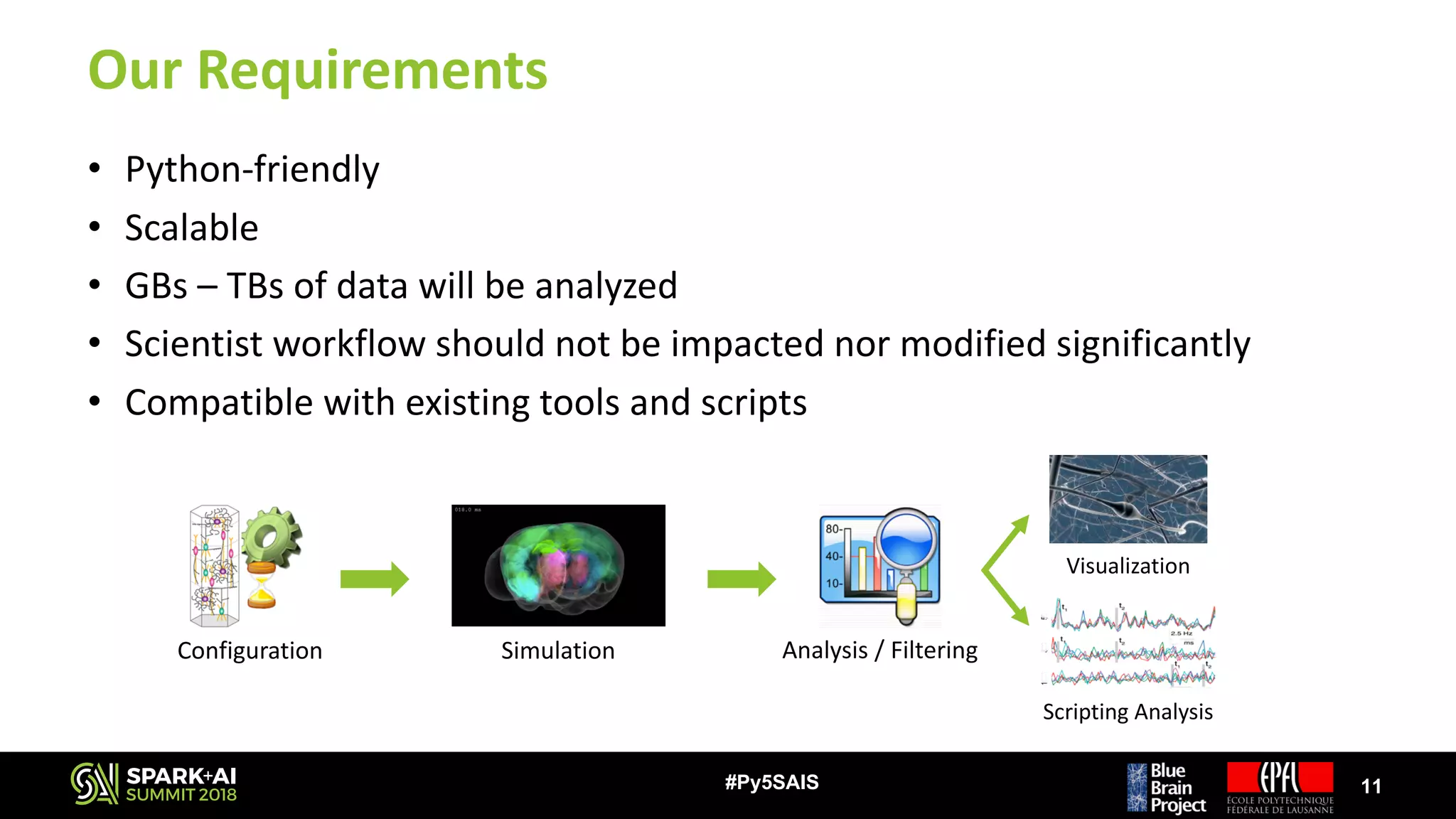
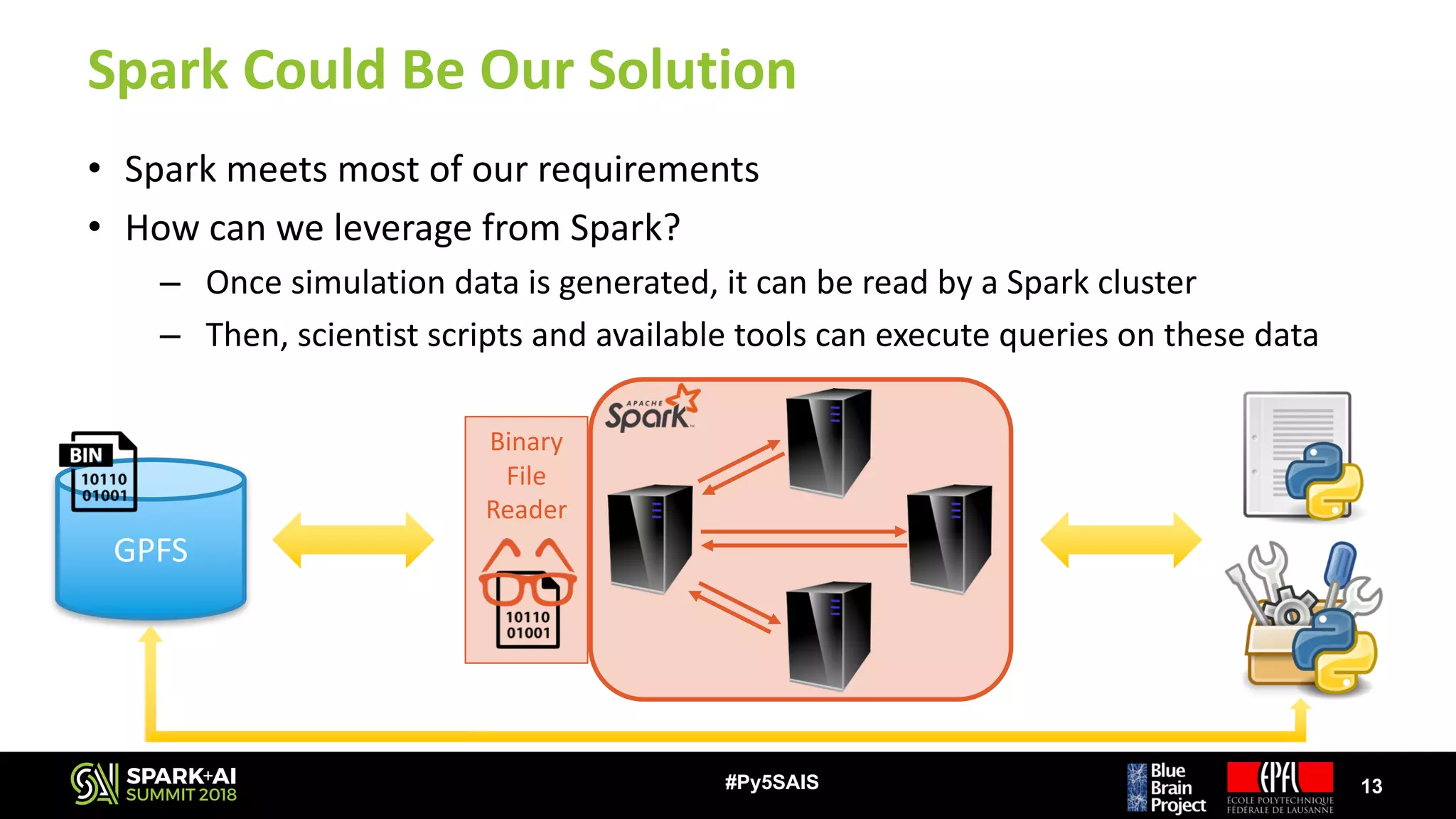
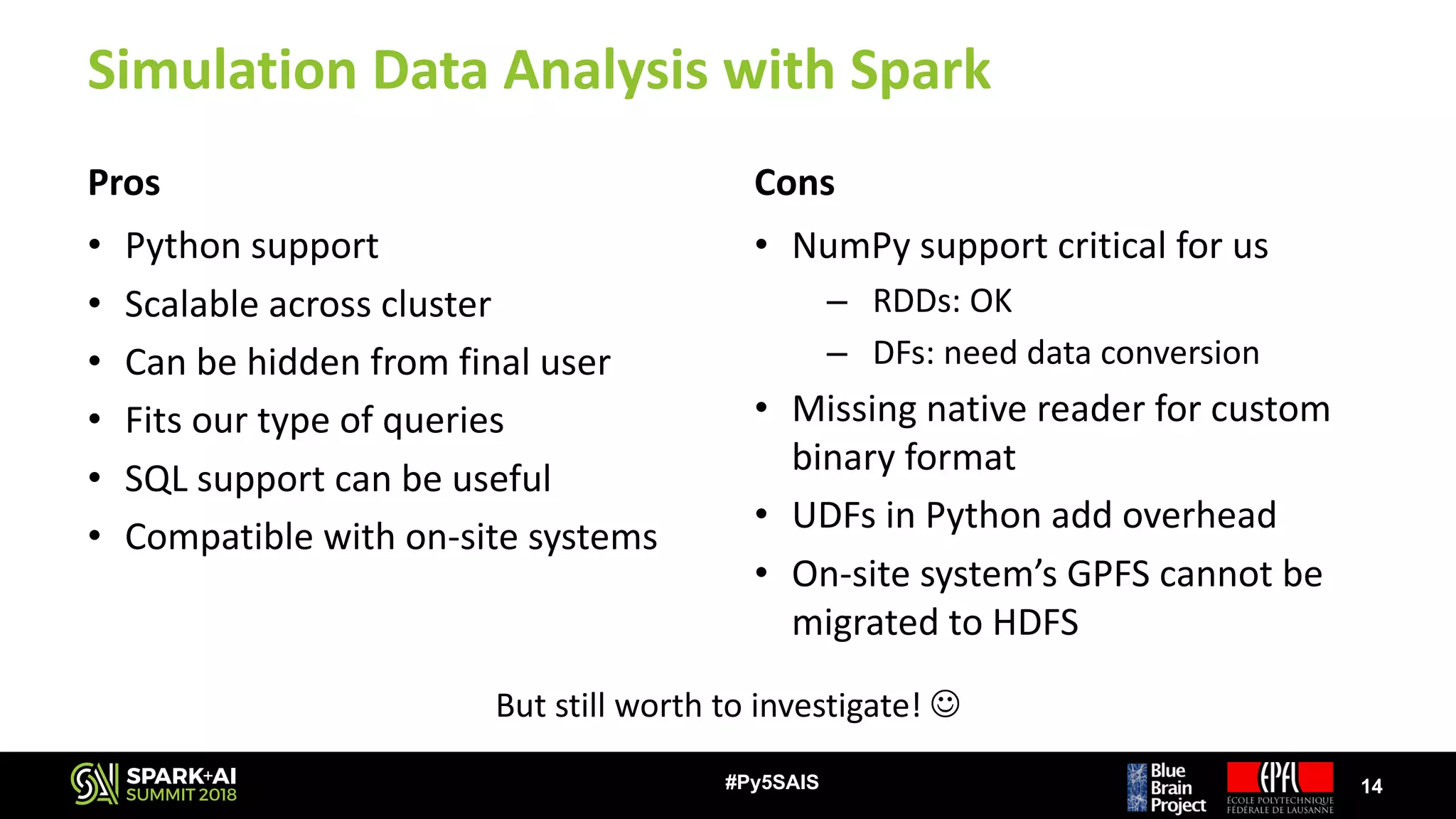
![Our Challenge
• Simulations can produce GBs – TBs of data very quickly
– E.g.: Plastic neocortex simulation
• Recording 1 variable
• 30 s of biological time ~50 GB of output data [ x N ]
• 31’000 neurons
⭐ But… >1 variables are recorded
⭐ And… Biological time is much longer
• Most scientists use sequential scripts to analyze their data
– Most preferred language is Python
– They do not have time / knowledge to improve their scripts
– Existing analysis tools exploit thread-level parallelism on single node
10#Py5SAIS](https://image.slidesharecdn.com/5juditplanas-180618234325/75/Accelerating-Data-Analysis-of-Brain-Tissue-Simulations-with-Apache-Spark-with-Judit-Planas-10-2048.jpg)

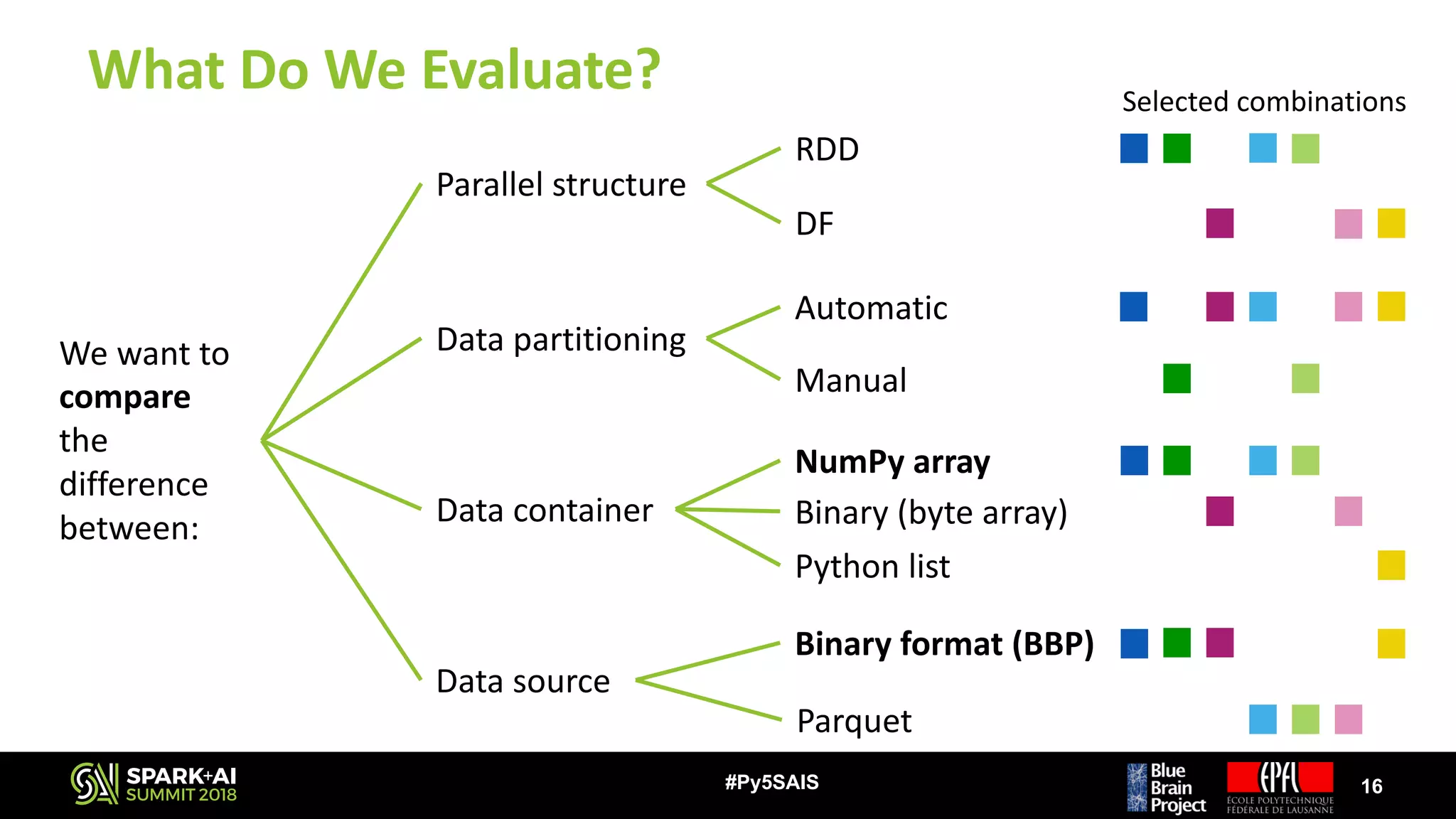
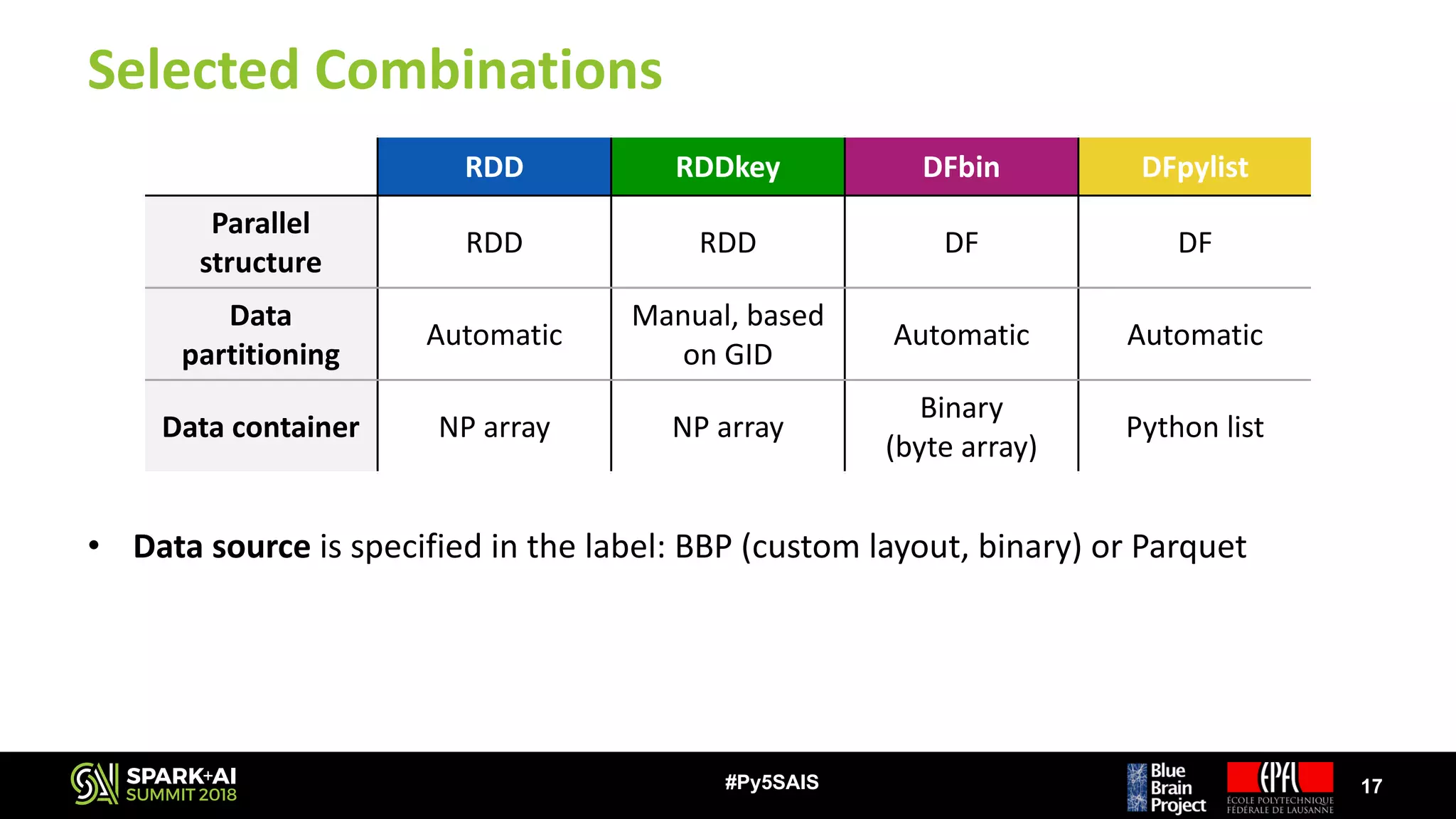
![Memory Footprint
20#Py5SAIS
0
50
100
150
200
250
300
Node Memory Footprint [GB]
DFpylist - BBP RDD - Parquet RDDkey - Parquet DFbin - BBP
DFbin - Parquet RDDkey - BBP RDD - BBP
• Original dataset size (files): 47 GB
• RDD – BBP is the only version that
stays close to original files size
• Any other data source / data structure
increases memory consumption from
3 – 6 times
• DFpylist crashes due to lack of
memory while performing the
computation by GID (unable to
continue with memory test)](https://image.slidesharecdn.com/5juditplanas-180618234325/75/Accelerating-Data-Analysis-of-Brain-Tissue-Simulations-with-Apache-Spark-with-Judit-Planas-20-2048.jpg)
![Data Loading Performance
• Loading data from original files (BBP)
does not scale: file system is too
overloaded accessing the same files
• Loading data from Parquet scales
because data is split into 1000s of
files, but also hits GPFS limitations
• RDDkey takes more time than the
others because of the extra
repartitioning of data
• DFbin takes more time than RDD due
to data conversion
21#Py5SAIS
0
1000
2000
3000
4000
5000
6000
7000
8000
8 16 24 32 40
Execution time [s]
# Nodes
RDD - BBP RDDkey - BBP DFbin - BBP
RDD - Parquet RDDkey - Parquet DFbin - Parquet
Existing tools (thread-level parallelism, 1
node): 13’820 s à up to 100x speed-up!](https://image.slidesharecdn.com/5juditplanas-180618234325/75/Accelerating-Data-Analysis-of-Brain-Tissue-Simulations-with-Apache-Spark-with-Judit-Planas-21-2048.jpg)
![Reduction by GID/TS Performance
22#Py5SAIS
0
100
200
300
400
500
600
700
800
900
8 16 24 32 40
Execution time [s]
# Nodes
Reduce by GID
RDD - BBP RDDkey - BBP DFbin - BBP
RDD - Parquet RDDkey - Parquet DFbin - Parquet
0
10
20
30
40
50
60
8 16 24 32 40
Execution time [s]
# Nodes
Reduce by TS
• RDDkey is the fastest thanks to the manual
partitioning: better data distribution
• DFbin is the slowest due to data conversions
• Reduce by TS was run right after Reduce by
GID: we believe that partial results were
cached and, thus, is significantly faster
Existing tools (TLP, 1 node): 5 s à worse
or same perf à needs investigation](https://image.slidesharecdn.com/5juditplanas-180618234325/75/Accelerating-Data-Analysis-of-Brain-Tissue-Simulations-with-Apache-Spark-with-Judit-Planas-22-2048.jpg)
![Computation by GID/TS Performance
23#Py5SAIS
0
50
100
150
200
250
300
350
400
8 16 24 32 40
Execution time [s]
# Nodes
Comp by GID
RDD - BBP RDDkey - BBP DFbin - BBP
RDD - Parquet RDDkey - Parquet DFbin - Parquet
0
20
40
60
80
100
120
140
160
180
200
8 16 24 32 40
Execution time [s]
# Nodes
Comp by TS
• RDDkey is again the fastest
• RDD Parquet needs the inverse data
conversions (from bin to NP)
• Comp by TS was run right after Comp by GID
• Significant speed-up from 8 to 16 nodes
(partially due to external load in the system)
Existing tools (TLP, 1 node):
2347 s à up to 130x speed-up!](https://image.slidesharecdn.com/5juditplanas-180618234325/75/Accelerating-Data-Analysis-of-Brain-Tissue-Simulations-with-Apache-Spark-with-Judit-Planas-23-2048.jpg)

![Acknowledgments & References
• BBP In Sillico Experiments and HPC teams for the support and feedback provided
• An award of computer time was provided by the ALCF Data Science Program (ADSP). This
research used resources of the Argonne Leadership Computing Facility, which is a DOE Office
of Science User Facility supported under Contract DE-AC02-06CH11357
[1] Henry Markram et al. Reconstruction and Simulation of Neocortical Microcircuitry. Cell, Vol.
163, Issue 2, pp 456 – 492
[2] Kunkel, Susanne et al. Spiking network simulation code for petascale computers. Frontiers in
neuroinformatics, Vol. 8, p 78
[3] Ovcharenko, A. et al. Simulating Morphologically Detailed Neuronal Networks at Extreme
Scale. In PARCO, pp. 787-796
[4] Chen, Weiliang et al. Parallel STEPS: Large Scale Stochastic Spatial Reaction-Diffusion
Simulation with High Performance Computers. Frontiers in neuroinformatics, Vol. 11, p 13
27#Py5SAIS](https://image.slidesharecdn.com/5juditplanas-180618234325/75/Accelerating-Data-Analysis-of-Brain-Tissue-Simulations-with-Apache-Spark-with-Judit-Planas-27-2048.jpg)
