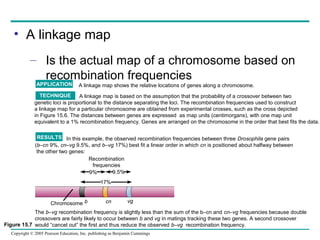
This document summarizes the chromosomal basis of inheritance and Mendelian genetics. It discusses how genes are located on chromosomes and can be visualized. It describes how Mendel's laws of segregation and independent assortment can be explained by the behavior of chromosomes during meiosis. Thomas Hunt Morgan provided evidence that genes are located on chromosomes through experiments with fruit flies. His work showed that linked genes on the same chromosome tend to be inherited together, while genes on different chromosomes assort independently. The document also discusses genetic recombination through crossing over and how this allows the construction of linkage maps. Finally, it describes different systems of sex determination and inheritance patterns of sex-linked genes.